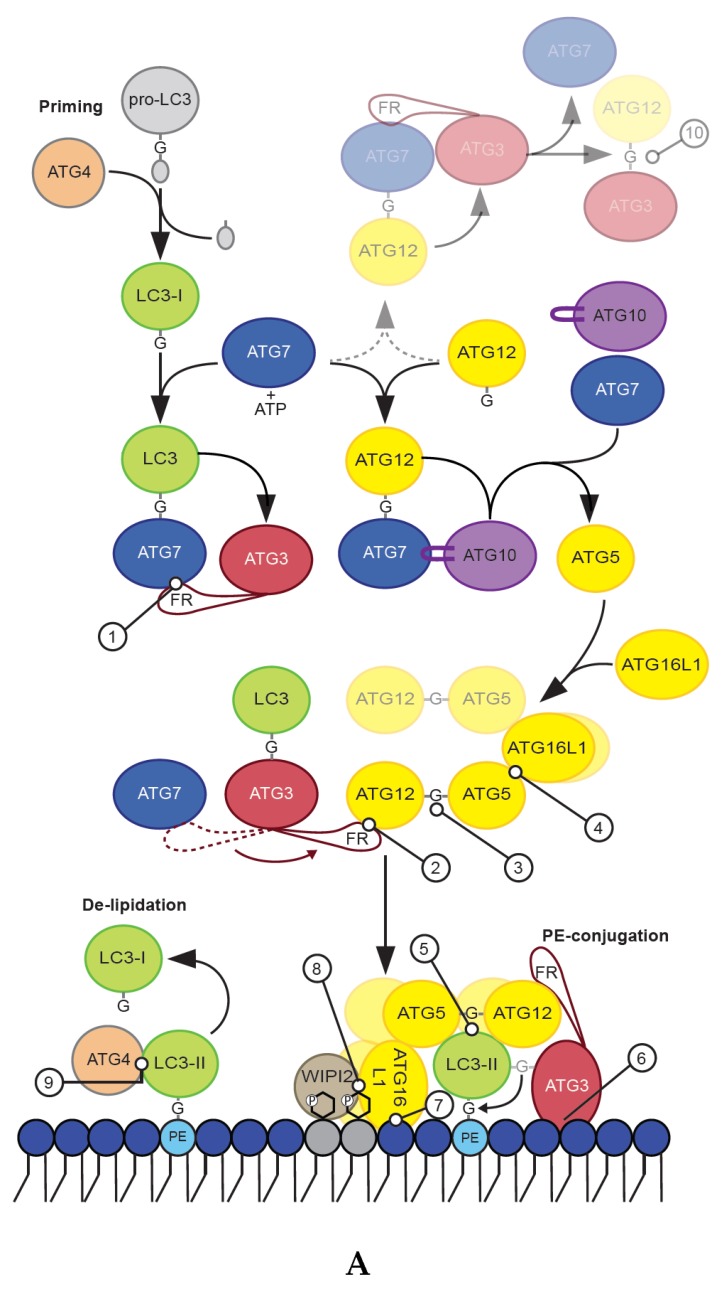
Figure 2.
(A) Mechanisms and interactions (protein and membrane) of core autophagy components required for priming, lipidation and de-lipidation of ATG8 proteins during starvation-induced autophagy are illustrated. The illustration also includes the conjugation of ATG12 to ATG5 as well as the less commonly reported conjugation of ATG12 to ATG3 (shaded), which serves its purpose outside of autophagy. Key interactions are numbered and shown in greater detail in (B). (B) ATG3 interacts with ATG7 and ATG12 through motifs located in a flexible region (aa 88–192), these motifs are referred to as RIA7 (1) and RIA12 (2) respectively. An amphipathic helix in the very N-terminal end of ATG3 (aa 1–26) provides membrane binding essential for lipidation of ATG8 proteins (6). G140 of ATG12 is conjugated to K130 of ATG5 (3) or K243 of ATG3 (10). It is proposed that V62 and W139 of ATG12 (5) serves as a LIR motif to secure the complex to the membrane of an autophagosome during lipidation. ATG16L1 interacts with ATG5 through its N-terminal helix (aa 13–28) (4). Membrane interaction of ATG16L1 is achieved through multiple interactions; an amphipathic helix near the N-terminal end provide membrane binding and is required for ATG8 lipidation (7), conserved residues in the coiled-coil domain together with its interaction with the phosphatidylinositol 3-phosphate (PtdIns(3)P) effector protein WIPI2 (8) provide binding to PtdIns(3)P and membrane binding through the β-isoform specific insert is important for ATG8 protein lipidation to damaged endosomes/lysosomes. The β-isoform specific insert is a highly post-translationally modified area in ATG16L1 and includes a confirmed phosphorylation site for ULK1 on S278. ATG4B interacts with ATG8 proteins through its catalytic core (CC) and an N-terminal and a C-terminal LC3-interacting region (LIR) (9).