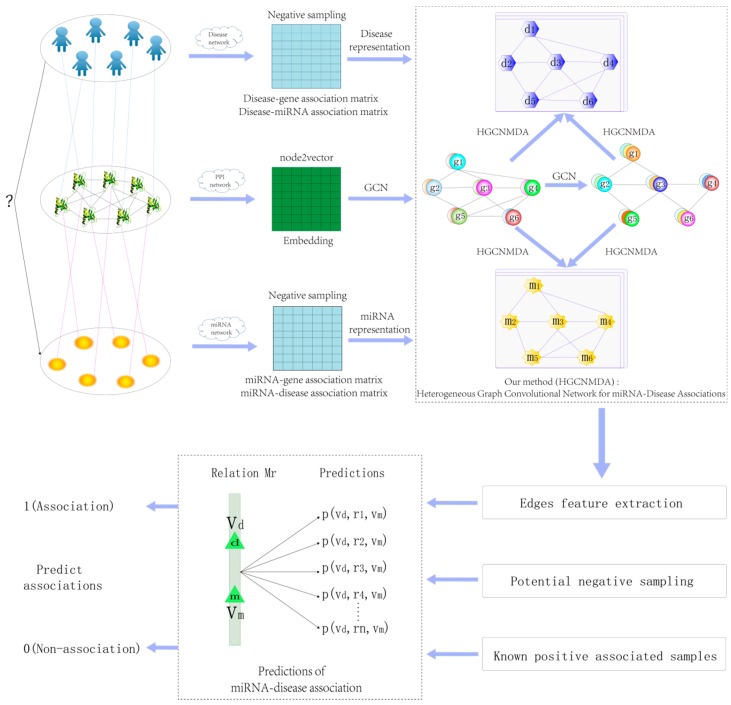
Figure 1.
The overall structure of heterogeneous graph convolutional network for miRNA–disease associations HGCNMDA. Input graphs of arbitrary structure are first passed through graphs convolution layers where node information is propagated between neighbors. First, features in gene–gene association network are obtained using graph convolution. Secondly, disease features and miRNAs features are extracted respectively based on PPI features using HGCNMDA approach that we proposed. Lastly, the edge features between diseases and miRNAs are extracted and passed to decoder layer to train a predictive model after obtaining potential negative sampling together with known positive associated samples. Features are visualized as colors.