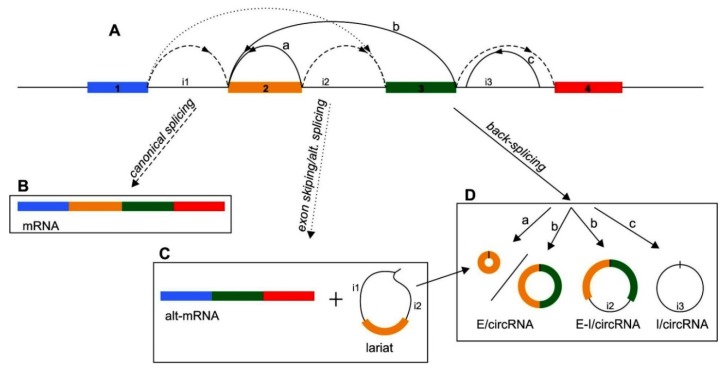
Figure 1.
Schematic presentation of circular RNAs (circRNAs) biogenesis. (A) Schematic representation of pre-mRNA with indicated exons (colored rectangles), introns (named as i1, i2 and i3), and different forms of splicing. Pre-mRNA undergoes a canonical (B) and non-canonical (C and D) splicing. (B) In a canonical splicing (dashed lines in panel A) consecutive exons are joined together (i.e., exon 1 to exon 2 to exon 3, etc.) to generate a linear mRNA that is subsequently translated. (C and D) Pre-mRNA may also be processed in a non-canonical splicing via either exon skipping (C), (dotted lines in panel A) or via back-splicing (D), (solid lines in panel A). Exon skipping events can lead to the generation of a linear mRNA when, e.g., exon 1 is joined directly to exon 3. It is worth noting that lariat is a byproduct of exon skipping, and may facilitate back-splicing, which eventually gives rise to a circRNA composed of the skipped exon (e.g., exon 2) (C). Different types of circRNAs (D), i.e., exonic (E/circRNAs), exonic-intronic (E-I/circRNA) and intronic (I/circRNA) are generated through back-splicing of the 5′ splice donor to the 3′ splice acceptor (e.g., joining the 3′ end of exon 3 to the 5′ end of exon 2). E/circRNAs may be generated from: (a) a single exon (e.g., exon 2), or from (b) two or more exons (e.g., exons 2 and 3). The E-I/circRNAs (b) may be generated from retaining intron which separates exons participating in circRNA formation (e.g., exons 2 and 3, and intron 2). However, the I/circRNAs (c) may be generated through interactions of reverse complementary sequences present in lariat introns (e.g., intron 3) excised from pre-mRNA.