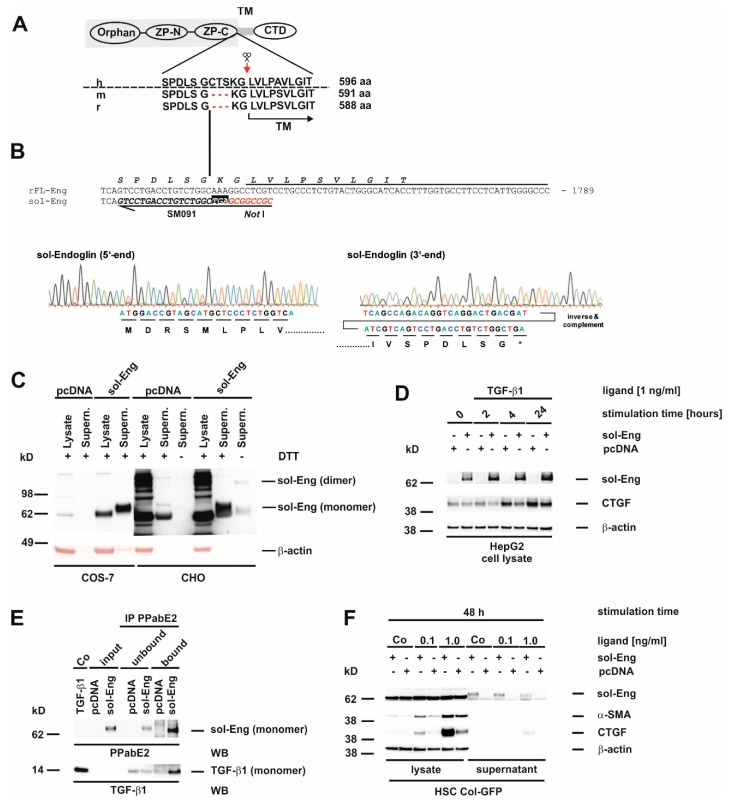
Figure 3.
Structure, sequence analysis and functional expression of soluble Endoglin from rat. (A) Scheme of the FL-Endoglin (FL-Eng) domain structure including the amino acid environment surrounding the cleavage site (scissors and red arrow) of human, mouse and rat. (B) Nucleotide sequence of FL-Eng and the corresponding primer composition (SM091) to amplify sol-Eng and introduce a stop codon (TGA) and a NotI restriction site (upper panel). Sequence analysis of the sol-Eng expression plasmid (lower panel). For clarity the sequence data of the 3′end was inversed and complemented for depicting the encoded amino acids. (C) COS-7 cells (left) and CHO cells (right) were transiently transfected with empty vector (pcDNA) or sol-Eng expression vector (sol-Eng). sol-Eng expression was displayed in Western blot in the presence (+) or absence (−) of DTT. (D–E) HepG2 cells were transiently transfected using empty vector (pcDNA) or sol-Eng expression vector (sol-Eng). Thereafter, cells were either treated or not with TGF-β1 (1 ng/mL) for the indicated time points and sol-Eng expression and CTGF expression was monitored in Western blot (D) or the supernatant was supplemented with TGF-β1. Binding was accomplished for 2 h rotating and sol-Eng complexes were precipitated overnight using a rat Eng-specific antibody (PPabE2) and analysed in Western blot for the presence of bound TGF-β1 and precipitated sol-Eng. (F) HSC Col-GFP cells were transiently transfected using empty vector (pcDNA) or sol-Eng expression vector (sol-Eng). After starvation for 16 h, the cells were stimulated or not for 48 h with TGF-β1 (0.1, 1.0 ng/mL). Cellular proteins were analysed in Western blot for transgene expression (sol-Eng), CTGF and α-SMA. Equal loading of proteins was proven by re-hybridization of the membranes with a β-actin antibody.