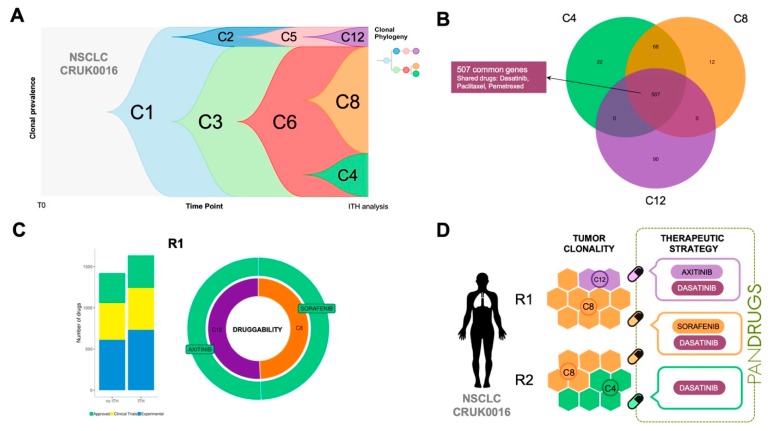
Figure 5.
Analysis of the non-small cell lung cancer (NSCLC) patient CRUK0016. Two regions (R1 and R2) were sampled and three different clones (C4, C8 and C12) were detected. (A) Fishplot and clonal phylogeny of the evolution of tumoural cell populations. (B) Venn diagram of the mutated genes detected in the tumour subclones. PanDrugs proposed dasatinib, paclitaxel and pemetrexed to target the common genes among all clones (trunk). (C) Left: Number of therapeutic candidates revealed by bulk and intra-tumour Heterogeneity (ITH) analysis of R1. We assumed that bulk sequencing would only detect the predominant clone (C8). Drugs have been coloured according to their status. Right: Druggability and proposed therapies to treat the clones in R1. Druggability is defined as the percentage of different targetable genes in each subclone over the total targetable genes in a region. (D) Therapeutic regimen proposed to treat patient CRUK0016 based on PanDrugs predictions. Dasatinib could be used to target all clones simultaneously (trunk), while axitinib and sorafenib would target clone-specific mutations in C12 and C8 respectively. We did not find any good drug candidate (Drug Score > 0.70) to treat C4 individually. Thus, R1 could be treated with a combination of drugs targeting trunk mutations (dasatinib) and two subclone-specific drugs whereas R2 would be targeted with dasatinib and C8-specific treatment (sorafenib).