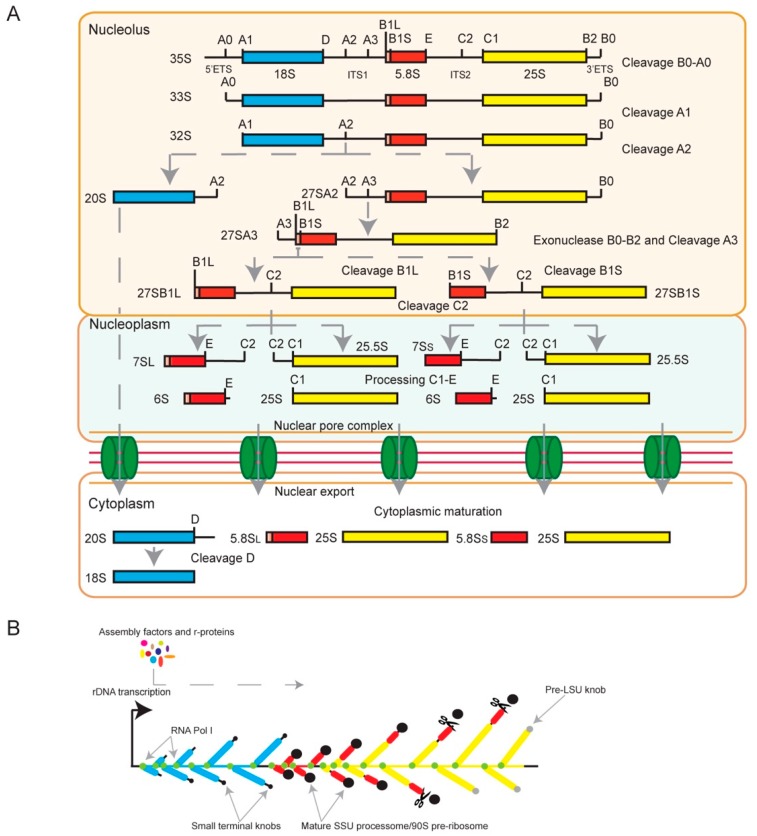
Figure 1.
Processing of pre-ribosomal RNA (pre-rRNA) and small subunit (SSU) processome formation in the yeast Saccharomyces cerevisiae. (A) Scheme of 35S pre-rRNA processing steps. The 35S precursor is cleaved and trimmed at different sites to form the mature 18S (blue), 5.8SL, 5.8SS (red) and 25S (yellow) rRNAs. Processing takes place in different cellular compartments (nucleolus, nucleoplasm and cytoplasm), represented by rectangles. Cleavage sites are indicated on the transcripts and are detailed in the text. The final maturation steps occur in the cytoplasm once the late precursors have been exported via the nuclear pore complex (NPC, colored in green). (B) Representation of SSU processome formation. The ribosomal DNA (rDNA) is transcribed by RNA polymerase I (Pol I) (green circles). When visualized by electron microscopy, actively transcribed rDNA units appear as Christmas trees, wherein chromatin forms the trunk, nascent rRNA transcripts are the branches, and the terminal knob seen at the 5′ end of transcripts is the forming SSU processome. The small black circles represent the newly formed SSU knobs and the larger black circles are the mature SSU knobs, where the 5′ETS and the 18S rRNA are packed. The large SSU knob is then cleaved at site A2 (represented by the black scissors). The large subunit precursor (pre-LSU) knobs are shown in grey circles. This schematic representation is adapted from Osheim et al. [70].