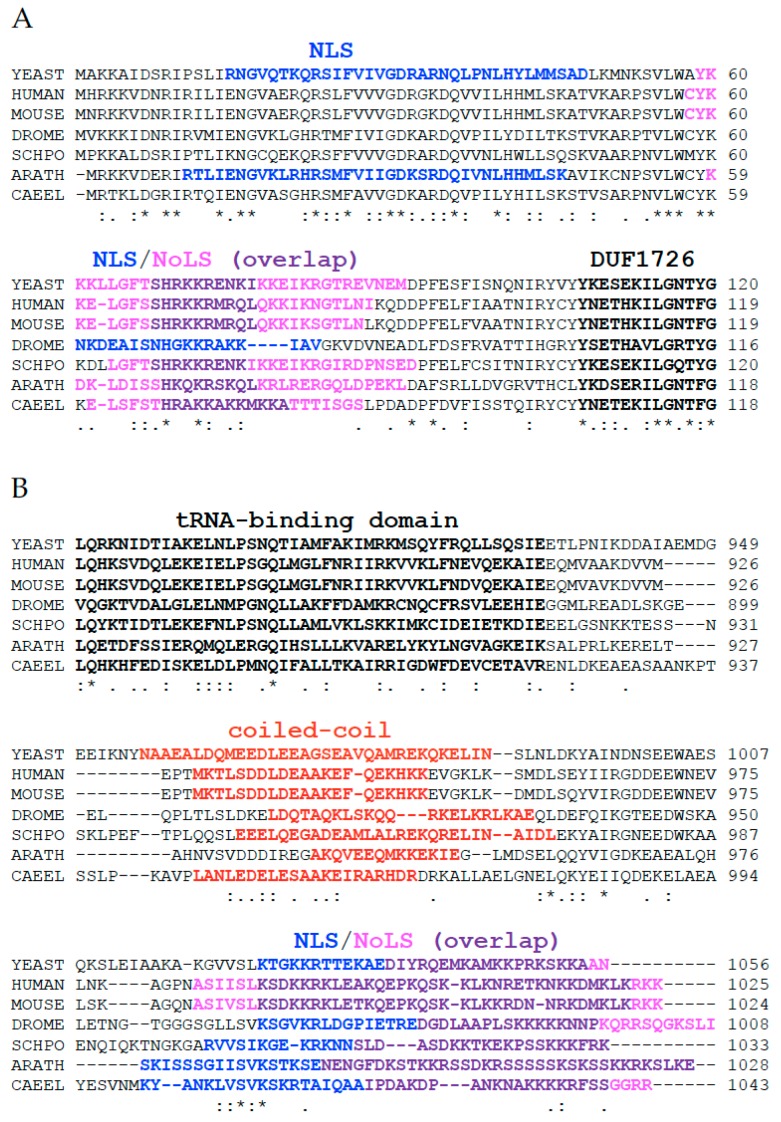
Figure 3.
Alignment of Kre33/NAT10 from different species. Sequences from yeast, human, mouse, D. melanogaster (DROME), S. pombe (SCHPO), A. thaliana (ARATH) and C. elegans (CAEEL) were used to generate an alignment with Clustal Omega [102]. (A) The N-terminal extension precedes the DUF1726 (black bold letters) and harbors eukaryote-specific motifs, such as the NLS (blue) and the NoLS (magenta); overlapping NLS/NoLS sequences are in purple. (B) The C-terminal extension follows the tRNA-binding domain (black bold letters): it contains a coiled-coil motif (red) and finishes with the NLS (blue)/NoLS (magenta); the NLS/NoLS overlap is in purple. The putative NLS, NoLS and coiled-coil motif were identified with cNLS mapper [103], NoD [104] and COILS [105].