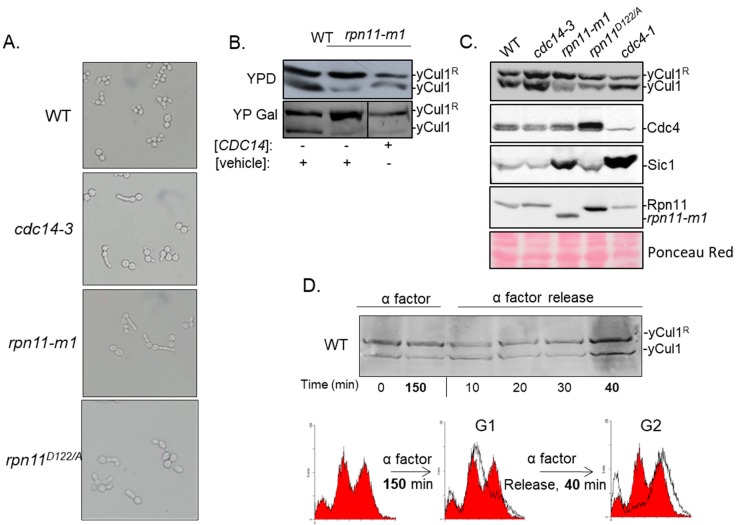
Figure 2.
Cell cycle does not determine yCul1 NEDDylation status. (A) WT and mutant yeast strains were grown at 28 °C for 16 h, then shifted to a restrictive temperature of 37 °C for 5 h to enhance cellular phenotypes. (A) Cell cycle defects were observed by light microscope (magnification x400). (B) The rpn11-m1 mutant cells were grown overnight in raffinose and diluted to 0.5 OD600 in galactose (YP Gal) to induce the expression of CDC14 or a vehicle plasmid, or in glucose (YPD) for control. The extent of yCul1 modification by Rub1 was examined by immunoblotting of total protein extracts with yCul1 antibody. (C) Yeast strains were grown as previously explained (A), followed by extraction of total protein extracts used for yCul1 immunoblotting. The accumulation of the short-lived F-box protein Cdc4 and its substrate Sic1 was validated as well. Note that cdc4-1 is a control for decreased endogenous levels of the yCul1Cdc4 complex. Cdc4 expression shows a negative correlation with the accumulated Sic1. (D) WT cells were grown in glucose supplemented by α factor for 150 min for synchronization of the cell cycle in G1. Synchronized cells were washed and were then grown in YPD. Samples were taken before and after synchronization at indicated times and subjected to immunoblotting with yCul1. The cell cycle stage was analyzed by quantitation of DNA content by flow cytometry.