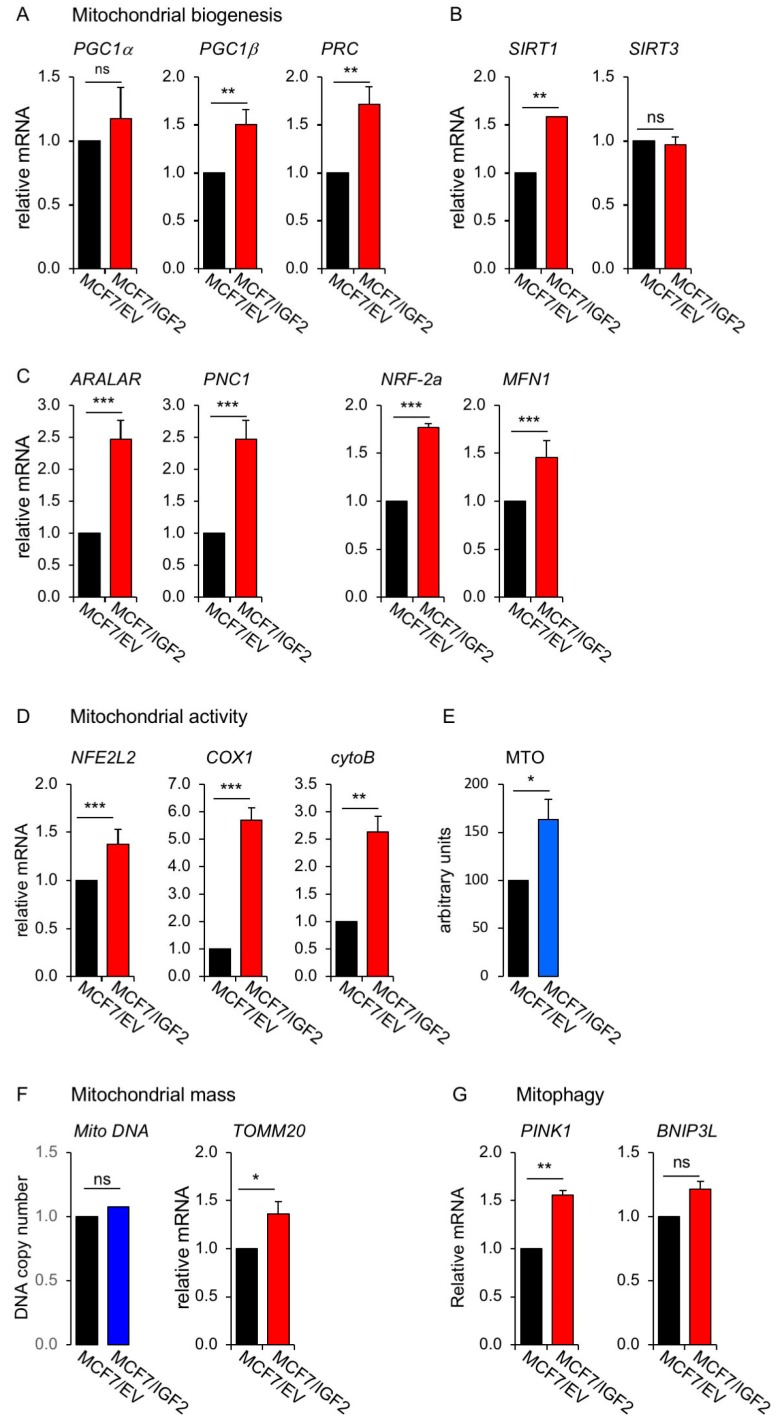
Figure 3.
Markers of mitochondrial biogenesis and activity, mitochondrial mass and mitophagy were regulated in MCF7/IGF2 cells. (A) mRNA expression levels of mitochondrial genes, peroxisome proliferator-activated receptor gamma coactivator-1 (PGC1)α, PGC1β and PGC1α-related coactivator (PRC); (B) Sirtuin (SIRT)1 and SIRT3 mRNA expression; (C) aspartate–glutamate mitochondrial carrier 1 (ARALAR), pyrimidine nucleotide carrier 1 (PNC1), nuclear respiratory factor (NRF)-2a, and mitofusin (MFN)1 mRNA expression. (D) mRNA expression levels of mitochondrial activity genes nuclear factor erythroid-derived 2-like-2 (NFE2L2), cytochrome C oxidase 1 (COX1) and cytochrome B (cytoB). (E) Mitochondrial activity was evaluated by fluorescence activated cell sorting (FACS) analysis using MitoTracker Orange (MTO) probe. (F) Mitochondrial mass was assessed by measuring mtDNA copy number (left panel), and mRNA levels of mitochondrial outer membrane (TOMM20) (right panel). (G) Mitophagy mediators: mRNA expression levels of phosphatase and tensin homolog (PTEN)-induced kinase 1 (PINK1) and B-cell lymphoma 2 (BCL2)/adenovirus E1B 19 kDa protein-interacting protein 3-like (BNIP3L). MCF7/IGF2 and MCF7/EV cells were cultured in 2.5% charcoal stripped-FBS medium, and mRNA levels were determined by qRT-PCR. MCF7/EV cells were used as calibrator and GAPDH used as the housekeeping control gene in all qRT-PCR experiments. Values are means ± SEM of three independent experiments. (ns, not significant; * p < 0.05; ** p < 0.01; *** p < 0.001).