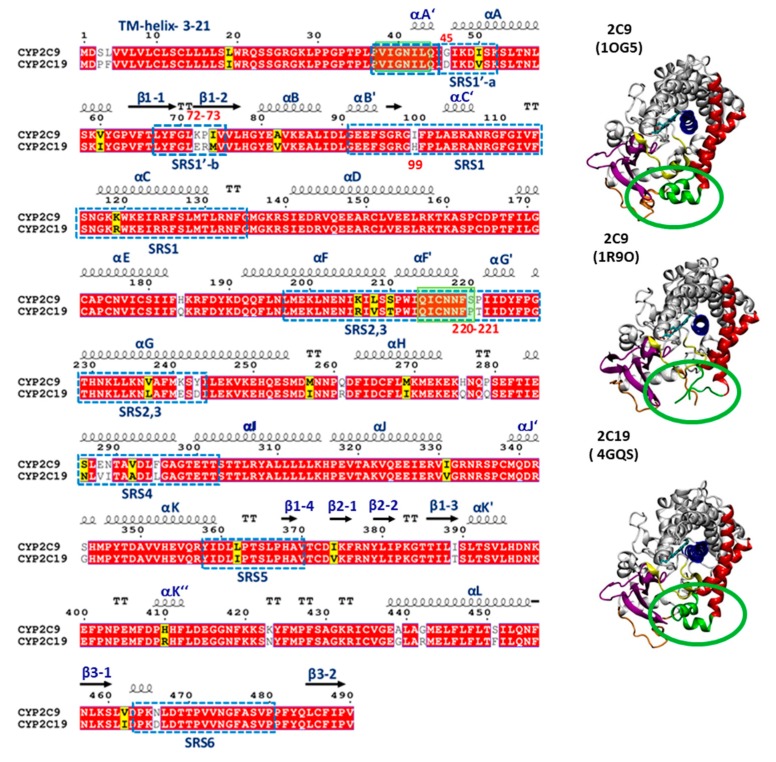
Figure 1.
(Left) Sequence alignment of CYP 2C9 and CYP 2C19. Identical residues are shown with a red background, similar residues with a yellow background, and differing residues with a white background. The secondary structure in the crystal structure of CYP 2C19 (PDB 4GQS) is indicated by arrows for β-strands, springs for α-helices, and ‘TT’ for turns; long loops are unmarked. The substrate recognition sites (SRS) are shown by blue dashed line boxes. The residues in the globular domain differing at the membrane interface are highlighted by red numbers. The missing regions in the crystal structure (PDB 1R9O) of the globular domain of CYP 2C9 are shown by transparent green boxes. (Right) Cartoon representations of the crystal structures of CYP 2C9m7 (PDB 1OG5), CYP 2C9 (PDB 1R9O), and CYP 2C19 (PDB 4GQS), showing the structural differences in the F’–G’ region highlighted by the green rings, the heme in stick representation, and key secondary structure elements colored as follows: β-strand regions in magenta, the B–C loop in yellow, the F and G helices in red, the F’–G’ helices/loop in green, the I-helix in blue, and the linker in orange. The active site is lined by the heme and the I-helix.