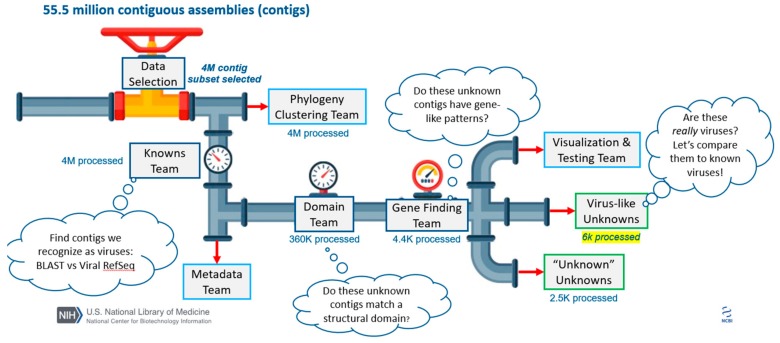
Figure 1.
Overview of hackathon teams and data processing. All numbers detail the number of contigs processed at each step of the pipeline. A subset of ~3000 data sets were assembled, generating 55.5 million total contigs. Researchers attending the hackathon assembled into teams that roughly correspond to goals outlined in the Methods and Results. Members of the “Knowns Team” excluded contigs based on size (removing <1 kb in length) and the remaining ~4 million contigs were assigned classification to known viruses using a BLASTN search against the RefSeq Virus database (Section 2.3 and Section 3.3). Independently, members of the “Phylogeny Clustering Team” clustered ~4 million contigs using Markov Clustering techniques (Section 2.4). Members of the “Metadata Team” used machine learning approaches to build training sets that could be used to correlate sequences to sample source metadata (Section 2.7 and Section 3.7). Members of the “Domain Team” predicted functional domains with RPSTBLASTN and the CDD database using ~360,000 contigs that were not classified using the RefSeq Virus database (Section 2.5 and Section 3.3). Members of the “Gene Finding Team” predicted open reading frames and putative viral-related genes using the modified VIGA pipeline on ~4400 putative viral contigs (Section 2.6 and Section 3.6). Members of the “Visualization Team” devised ways to display complex data and the “Testing Team” accessed if components of the pipeline were accessible to future users. Two additional teams were tasked with analyzing sequences, which could not be identified as confidently cellular or virus-like with the methods described above (Section 3.5).