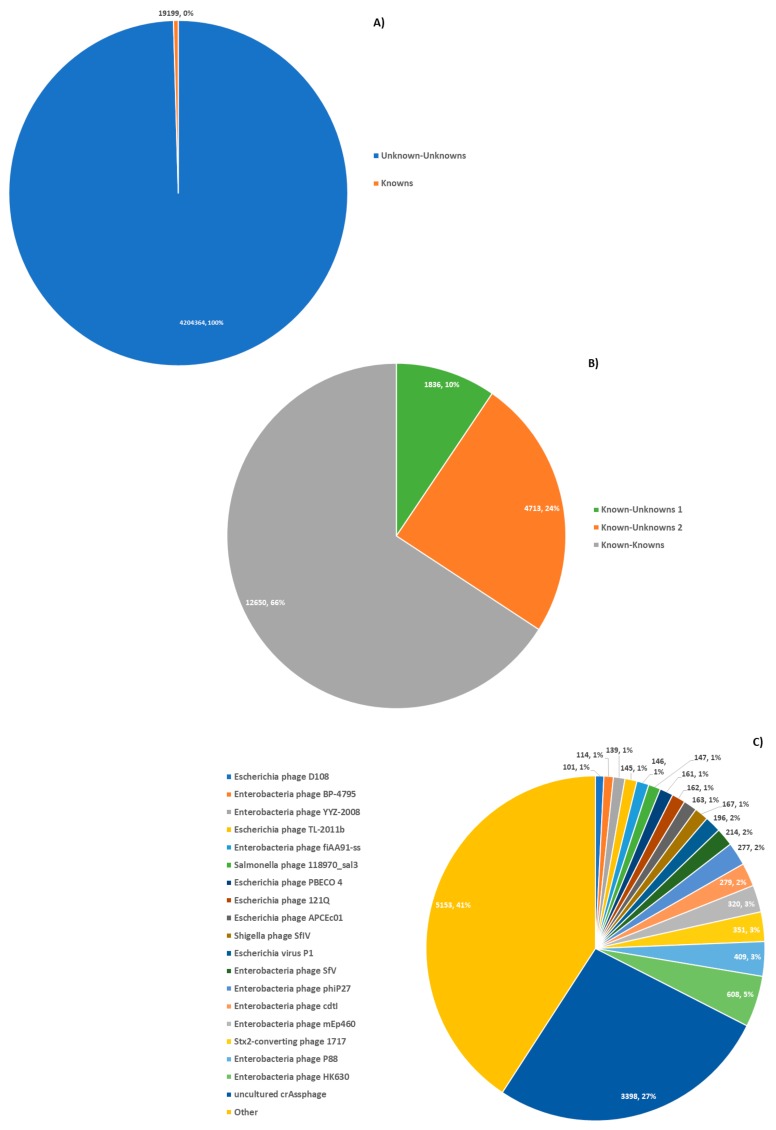
Figure 2.
Taxonomic classification of contiguous assemblies. Contiguous assemblies were compared to Virus RefSeq via BLASTN; (A) >99% of contigs have no hits amongst RefSeq viruses. (B) Of those with hits against RefSeq viruses, the majority had close matches, known–knowns. The remainder either had a weak match to a RefSeq, ‘known–unknowns’ 1, or a strong match to a RefSeq subsequence, ‘known–unknowns’ 2. (C) Top RefSeq viruses matched by ‘known–known’ contigs, by the number of contigs with a match to them. CrAssphage represents the most abundant virus in the subset.