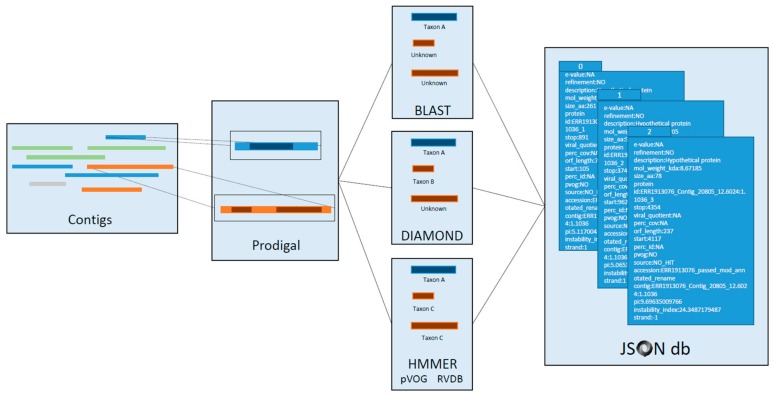
Figure 5.
Sequence annotation with VIGA. Putative viral sequences were annotated using the modified VIGA pipeline. Contigs depicting (putative) viral sequences, prefiltered by the presence of a viral CDD, were passed to VIGA. Each contig had their open reading frames (ORF) detected and translated with Prodigal using the 11th genetic code. Each viral protein was further characterized with a combination of three different annotation methods: BLAST, DIAMOND, and HMMER. HMMER included two model databases: pVOGs (prokaryotic viruses) and RVDB (all viral sequences but prokaryotic viruses). Coordinates, protein translation, hits, E-values, viral quotient, percentage of identity, and other meaningful information are codified in a hierarchical JSON database for downstream analysis.