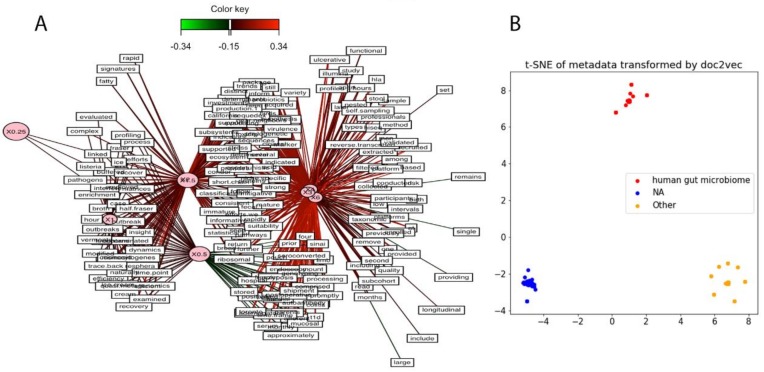
Figure 6.
Next generation sequencing (NGS) classification using associated metadata. (A) Study abstract and metadata are insufficient for NGS classification. Data sets where clustered using MASH, and partial least squares regression was performed to identify any covariance between the sequence content and word frequencies derived from the associated study abstracts and metadata; and (B) human gut microbiome samples are separable from other studies using Sequence Read Archive (SRA) metadata. Metadata was used as input to a word2vec model with 300 features, and the model was reduced to two dimensions using t-distributed stochastic neighbor embedding.