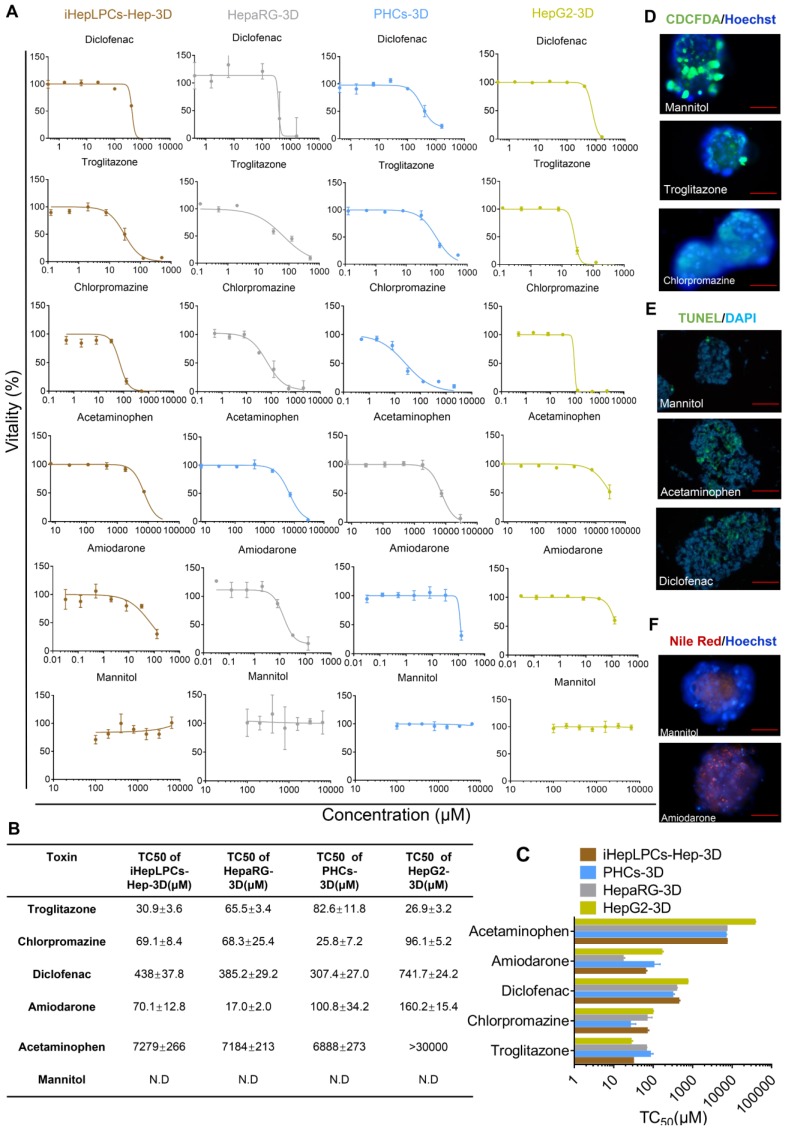
Figure 5.
iHepLPCs-Hep-3D can predict toxicological outcomes. (A) Dose-dependent toxicity curves of different compounds obtained from 48-h dose responses in iHepLPCs-Hep-3D, HepaRG-3D, hepG2-3D and PHCs-3D. (B-C) TC50 values of different compounds obtained from 48-h dose responses in iHepLPCs-Hep-3D, HepaRG-3D, hepG2-3D and PHCs-3D. Error bars represent s.d.; n = 3. (D-F) Fluorescence analysis of adverse outcome pathway in iHepLPCs-Hep-3D. Loss of bile acid production (cholestasis) evaluated by CDFDA staining, lipid accumulation (steatosis) by Nile Red staining and apoptosis by TUNEL labeling of nuclei. (D) Cholestasis in iHepLPCs-Hep-3D exposed to Troglitazone for 48 h, Chlorpromazine or Mannitol (negative control). (E) Apoptosis of differentiated hepatocytes following 48 h of exposure to Acetaminophen, Diclofenac or Mannitol. (F) Steatosis in iHepLPCs-Hep-3D after 48 h of exposure to Amiodarone or Mannitol. All error bars indicate ± s.d. All scale bar is 50 μm.