Figure 1. RhoA effector PKN1 mediates androgen-regulation of SRF target genes.
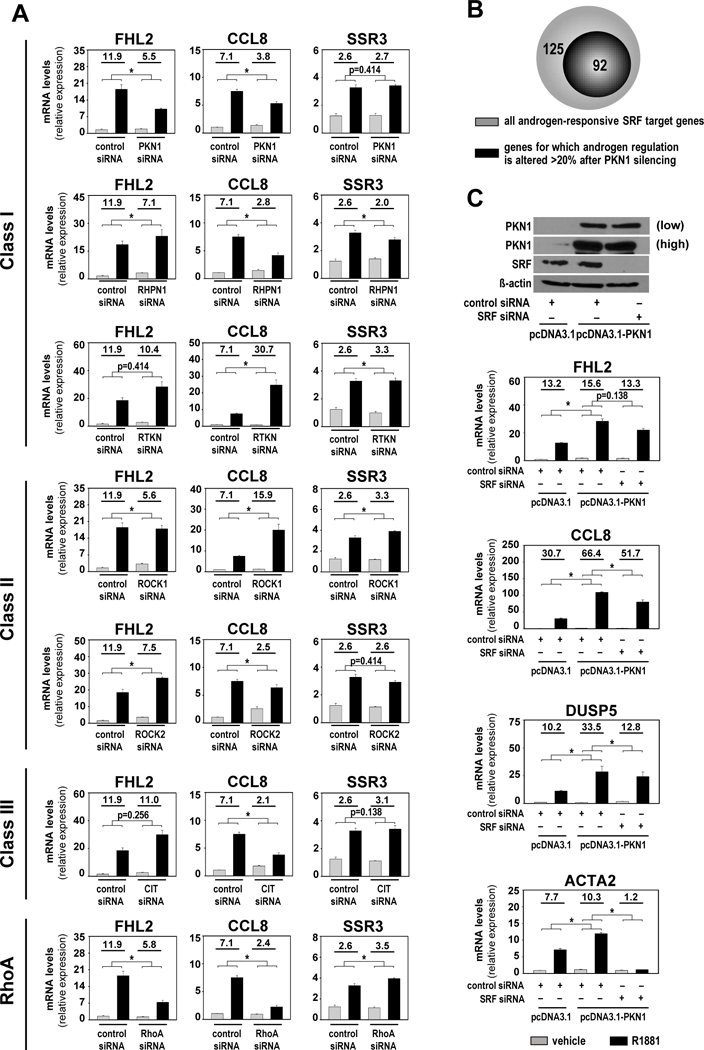
A. LNCaP cells were transfected with siRNA targeting RhoA or 6 RhoA effectors, or with non-specific control siRNA. At 42 hours after transfection, cells were treated with vehicle (ethanol) or 5nM of the synthetic androgen R1881. Cells were harvested 48 hours after treatment and androgen-induction of SRF target genes FHL2, CCL8 and SSR3 was evaluated via real-time RT-PCR. Target gene mRNA levels were normalized to GAPDH expression and are represented as relative expression using one of the values obtained from vehicle-treated control siRNA-transfected conditions as 1. Values obtained for specific siRNA transfections were evaluated against those from the same control siRNA group samples; all samples were from the same experiment. RhoA effectors Class I: PKN1, RHPN1, RTKN; Class II: ROCK1, ROCK2 and Class III: CIT. B. RNA-Seq was done on LNCaP cells after transfection with siRNAs targeting SRF or PKN1 or non-targeting siRNAs and treatment with 5nM R1881 or vehicle, using biological triplicates per treatment group as above. Genes that were ≥ 2-fold androgen-regulated (p adj <0.05) in control siRNA condition and for which androgen-responsiveness was decreased ≥1.5-fold upon loss of SRF were cross matched with gene sets that are bound directly by SRF. This approach isolated 217 androgen-responsive direct SRF target genes. Androgen regulation of 92 of these genes was affected >20% after PKN1 silencing. C. Bottom panel LNCaP cells were transfected with expression constructs encoding PKN1 (pcDNA3.1-PKN1) or empty vector (pcDNA3.1), in combination with control or SRF-targeting siRNA. At 42 hours after transfection, cells were treated with vehicle (ethanol) or 5nM R1881 and then harvested 48 hours after treatment. Androgen-induction of FHL2, CCL8, DUSP5 and ACTA2 was evaluated by real-time RT-PCR. Top panel Western blot analysis in a parallel experiment confirming the efficiency of PKN1 overexpression and siRNA-mediated knockdown of SRF. Blots were reprobed also for ß-actin as a loading control.
Columns, means of values obtained from independent biological triplicates; grey, vehicle treatment; black, R1881 treatment; bars, SEM values.*, p<0.05, all statistical analyses used Wilcoxon tests. Numeric values indicate the average fold change in androgen regulation. Low, low exposure, high, high exposure.
