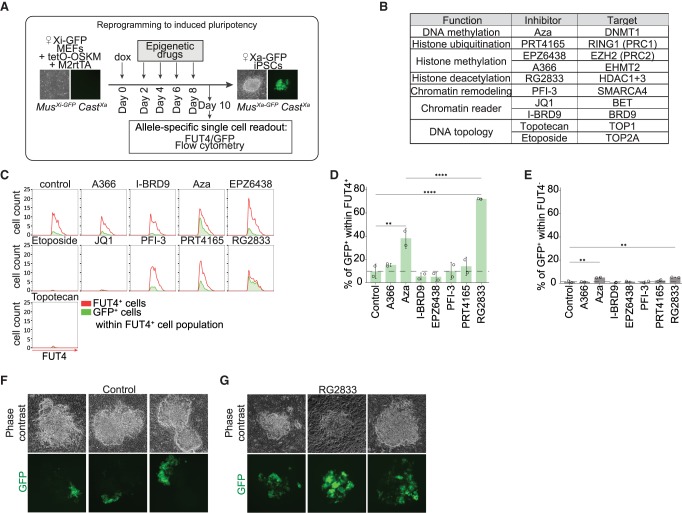
Figure 4.
Histone deacetylases restrict XCR during reprogramming to iPSCs. (A) A schematic representing experimental design of epigenetic drug inhibitor screen during reprogramming. (B) Inhibitors added individually at different time points during reprogramming with their function, name, and target molecule(s). (C) Histograms representing the flow cytometry analysis of the proportion of GFP+ cells within FUT4+ cells for each individual inhibitor at day 10 of reprogramming and vehicle control. The whole FUT4+ cell population is represented in red; population of GFP+ cells within FUT4+ cells, in green. (D) Proportion of GFP+ cells within the FUT4+ cell population for each individual inhibitor. The P-values of one-way ANOVA with Dunnett's multiple comparisons test comparing levels of GFP+ cells after treatment with each individual inhibitor and vehicle control are indicated with asterisks above the violin plots. The P-values of one-way ANOVA with Sidak's multiple comparisons test comparing levels of GFP+ cells treated with RG2833 and Aza are indicated with asterisks above the boxplot. (****) P-value < 0.0001; (***) P-value = 0.0001–0.001; (**) P-value = 0.001–0.01; (*) P-value = 0.01–0.05 = significant; (not significant) P-value ≥ 0.05. Error bars, SD. n = 2. (E) Proportion of GFP+ cells within FUT4− cell population for each individual inhibitor. The P-values of one-way ANOVA with Dunnett's multiple comparisons test comparing levels of GFP− cells after treatment with each individual drug inhibitor and vehicle control are indicated with asterisks above the boxplot. Error bars, SD. n = 2. (F) Phase contrast and fluorescent images of three representative colonies at day 10 of reprogramming after treatment with DMSO control during epigenetic drug screening. (G) Phase contrast and fluorescent images of three representative colonies at day 10 of reprogramming after treatment with RG2833, an inhibitor of HDAC1/3, during epigenetic drug screening.