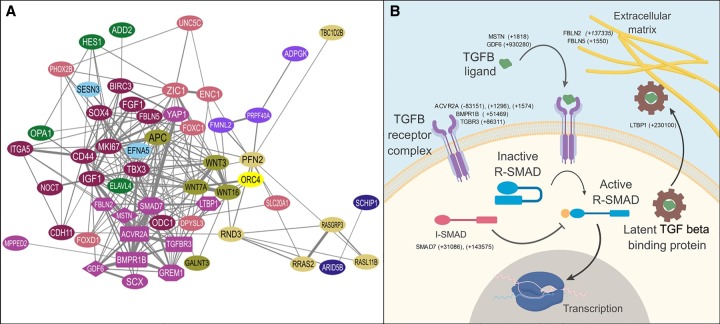
Figure 2.
Genes involved in TGF beta signaling are enriched among TWAR-associated genes. (A) STRING functional protein association network of TWAR-associated, TGF beta/BMP signaling, and responsive genes. Edges represent predicted interactions based on data contained with the STRING database for Mus musculus proteins. Node colors represent cluster membership based on MCL clustering. The purple (“violet blue”) network represents genes belonging to the TGF beta and BMP signaling pathways. In this cluster, ligands are shown as diamonds, receptors as rectangles, inhibitors as octagons, and proteins affecting ECM bioavailability of ligands as hexagons. Other clusters are composed of genes whose expression is known to be regulated by TGF-beta/BMP signaling; for example the small green-yellow (“highball”) colored cluster contains WNT signaling genes, whereas the dull yellow (“sandwisp”) cluster is composed of genes related to signal transduction by RAS superfamily GTPases. (B) Diagram outlining major components of TGF beta/BMP signaling pathways and TWAR-associated genes in each class. TWAR-associated genes of each type are given, with distances between TWARs and the transcription start site (TSS) of each gene given in parentheses. Minus indicates distance upstream (5′); plus, downstream (3′).