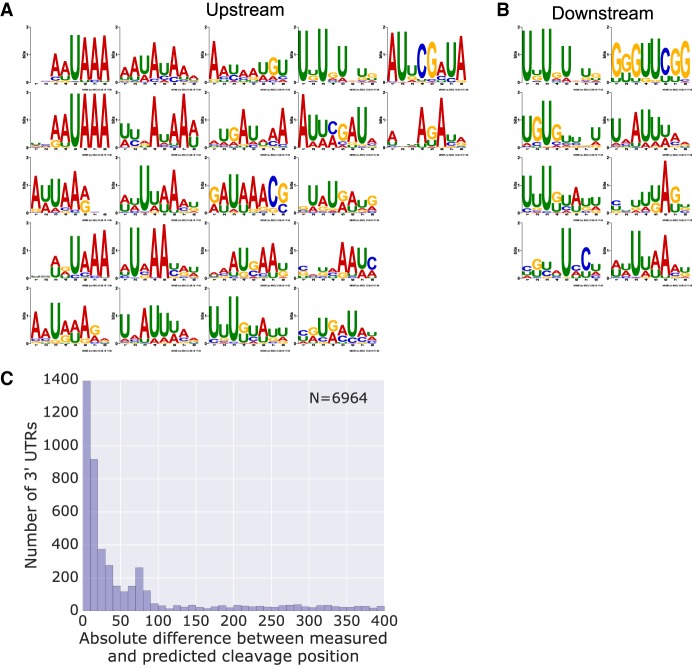
Figure 5.
The expression model learns biologically relevant sequence motifs which contribute to highly accurate endogenous cleavage site prediction. (A,B) Motifs were constructed for each filter from first layer activations (Methods). The motifs were analyzed for enrichment upstream of (A) and downstream from (B) endogenous cleavage sites using AME. Only motifs with an enrichment P-value < 0.001 are presented. (C) Histogram of the absolute differences between the measured and the predicted cleavage site on a set of endogenous 3′ UTRs (Methods). Predictions were made by applying the expression model on windows of 250-bp sequences shifted by 1 bp at a time. The position at which the maximal expression was achieved was adjusted by 145, the most likely cleavage position within the reporter library. See also Supplemental Figure S4.