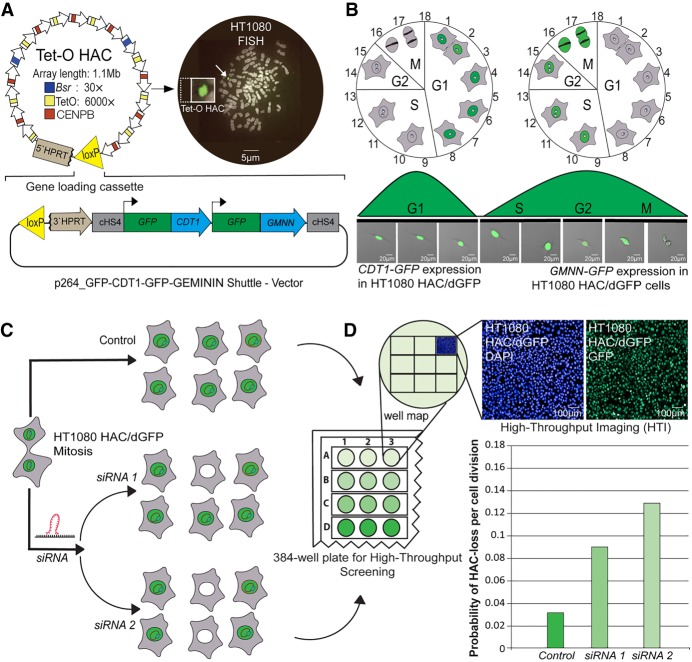
Figure 1.
Experimental design of a high-throughput imaging (HTI) human artificial chromosome (HAC)-based assay to identify novel CIN genes via siRNA screening. (A) The assay is based on the use of alphoidtetO-HAC (Tet-O HAC) (Nakano et al. 2008) expressing a dual short half-life green fluorescent protein GFP-CDT1-GFP-GMNN. Such HAC was named as HAC/dGFP. DNA in situ fluorescence hybridization (FISH) on a metaphase spread of HT1080 cells carrying HAC/dGFP using a probe against the HAC (Methods) and a schematic representation of the HAC/dGFP are shown. (B) A schematic representation of the dGFP reporter stability during different phases of the cell cycle in HT1080 cells. Cells that express cell cycle sensors CDT1 and GMNN fused with GFP display green fluorescence during the entire cell cycle. (C,D) Schematics of siRNA screening using HAC/dGFP-HTI assay to identify CIN genes. HT1080 cells carrying HAC/dGFP are transfected with a nontargeting control siRNA or with siRNA against a gene of interest, knockdown of which induces HAC/dGFP loss (indicated as siRNA1 or siRNA2), in a 384-well imaging plate. Cells are fixed, and the nuclear fluorescence in the GFP channel is measured using HTI. Cells transfected with siRNA against CIN genes display an increase of HAC/dGFP loss compared to the negative (nontargeting) control siRNA treatment.