Figure 2.
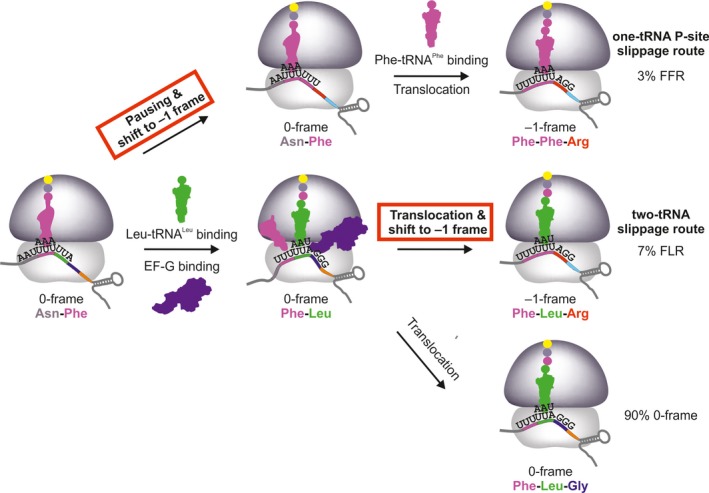
Kinetic mechanisms of FFR (upper) and FLR (lower) –1PRF pathways on the gag‐pol mRNA of HIV‐1. FFR results from one‐tRNA slippage with peptidyl‐tRNAP he in the P site (in magenta) when the A site is vacant due to low availability of Leu‐tRNAL eu( UAA ). FLR arises upon frameshifting during translocation of tRNAP he and peptidyl‐Phe‐Leu‐tRNAL eu( UAA ) (in green) and is prevalent at excess of Leu‐tRNAL eu( UAA ). After reading the slippery site, translation can continue in the –1‐frame by incorporating Arg at the AGG codon (red) or in 0‐frame by decoding Gly at the GGG codon (blue). The –1‐frame commitment steps on the FFR and FLR routes is marked in red. –1PRF on SFV 6K, IBV 1a/1b, and E. coli dnaX can, in principle, follow the same two routes. The existence of the two‐tRNA route is well‐documented 15, 29, 30, 45, 46, 57. The prevalence of the one‐tRNA route for SFV 6K depends on the concentration of tRNAL eu( UAA ) in the infected neuronal cells, which is not known (see text below). The one‐tRNA slippage on IBV 1a/1b could occur before decoding of the first slippery seqence codon UUA by the respective rare tRNAL eu( UAA ), however, the existence of the respective –1‐frame peptide product containing Phe‐Lys, rather than Leu‐Lys, has not been tested and the abundance of tRNAL eu( UAA ) in avian host cells is unknown. For dnaX, tRNAL ys that reads the slippery site codons is abundant and the one‐tRNA frameshifting pathway is only elicited by starvation.
