Figure 3.
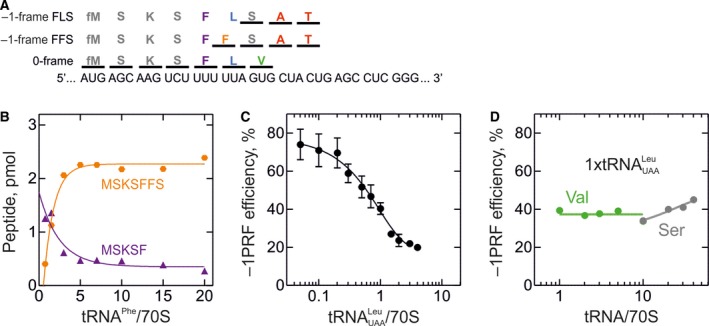
Mechanism of –1PRF on the SFV 6K mRNA. Translation was carried out in HiFi buffer at 37 °C as described in 69 for the gag‐pol mRNA; concentrations were Ser‐tRNAS er and Phe‐tRNAP he (with 0.8 μm each) and Lys‐tRNAL ys, Val‐tRNAV al, Ala‐tRNAA la and Thr‐tRNAT hr (0.25 μm each) and IC (0.08 μm) programmed with the 6K mRNA. Translation products were separated by reversed phase high‐performance liquid chromatography 69. 0‐frame products were identified based on the incorporation of [14C]Val, –1‐frame peptides using [14C]Ala and [14C]Thr. The –1PRF efficiency was calculated as a ratio between –1‐frame peptides and the sum of –1‐frame and all 0‐frame products, multiplied by 100%. (A) Schematic of the frameshifting site. The model SFV mRNA containing native SS and SL is optimized for translation in E. coli by introducing a SD sequence and a start codon AUG followed by AAG (Lys) to improve translation efficiency. (B) Effect of Phe‐tRNAP he on FFS peptide formation in the absence of Leu‐tRNAL eu( UAA ). Translation was carried out using tRNAs aminoacylated with M, S, K, F. (C) Dependence of –1PRF on Leu‐tRNAL eu( UAA ) concentration. Translation was carried out with M, S, K, F, L, V, A, and T aa‐tRNAs. (D) Effect of Val‐tRNAV al (green circles) and Ser‐tRNAS er (gray circles) concentrations on –1PRF efficiency. Translation was carried out using an equimolar concentrations of Leu‐tRNAL eu( UAA ) and aa‐tRNAs as in C. The large excess of Ser‐tRNA is required to ensure efficient translation of the SFV mRNA, which contains three Ser codons read by different tRNAS er isoacceptors in the total tRNAS er used in these experiments.
