Figure 1.
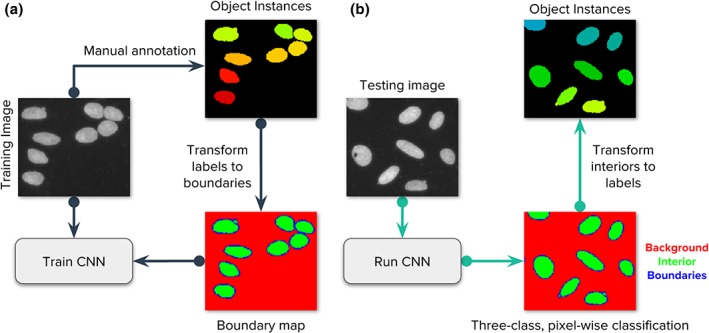
Strategy of the evaluated deep learning approaches. Our main goal is to follow the popular strategy of segmenting each nucleus and micronucleus as a distinct entity, regardless of whether it shares the same cell body with another nucleus. It is generally possible to group nuclei within a single cell using other channels of information in a postprocessing step if the assay requires it. (a) Images of the DNA channel are manually annotated, labeling each nucleus as a separate object. Then, labeled instances are transformed to masks for background, nucleus interior, and boundaries. A convolutional neural network (CNN) is trained using the images and their corresponding masks. (b) The trained CNN generates predictions for the three class classification problem. Each pixel belongs to only one of the three categories. In postprocessing, the predicted boundary mask is used to identify each individual instance of a nucleus. [Color figure can be viewed at wileyonlinelibrary.com]
