Figure 2.
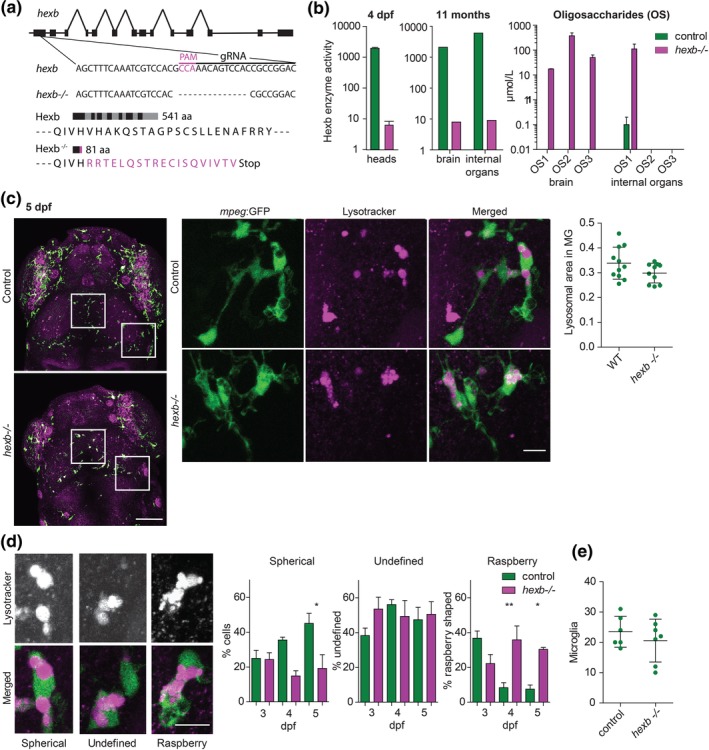
Stable hexb −/− fish show impaired Hexb enzyme function and lysosomal abnormalities in microglia. (a) Schematic representation of the hexb gene and the 18 bp gRNA and PAM motif. The hexb −/− fish show a 14 bp out of frame deletion disrupting part of the gRNA target sequence and PAM motif, resulting in 19 out of frame amino acids (magenta) followed by a premature stop codon. (b) Enzymatic substrate conversion by β‐Hexosaminidase A + B in larval heads and adult brain and internal organs—containing: intestine, liver, spleen, pancreas and gonads. Mass spectrometry analysis of Hexb deficiency associated oligosaccharide (OS) marker abundance based on exact mass (Hex2‐HexNAc2, 747.267 (OS1); Hex3‐HexNAc3, 1,112.399 (OS2); Hex3‐HexNAc4, 1,315.479 (OS3)) in 1.5 year old hexb −/− and control brains and internal organs—containing: intestine, liver, spleen, pancreas and gonads. (c) Representative images of microglia in LT‐stained larvae at 5 dpf, scale bar represents 100 μm. Quantifications were performed on detailed images of depicted regions, scale bar represents 10 μm. Quantification of relative lysotracker area within microglia. (d) Classification of lysosomal morphology in microglia with their quantification at 3, 4, and 5 dpf. n = 4 to 6 larvae per group. Scale bars represent 10 μm. (e) Quantification of total microglia in the imaged regions. Error bars represent SD. Each dot represents 1 larva [Color figure can be viewed at wileyonlinelibrary.com]
