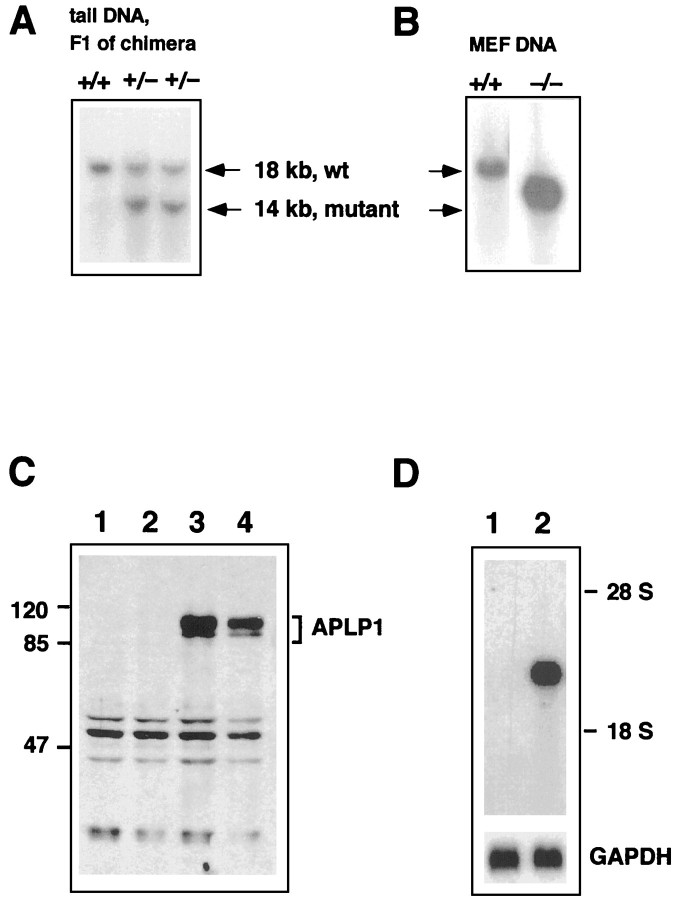
Fig. 2.
Molecular validation of the APLP1 knock-out. Functional inactivation of the APLP1 gene was confirmed by a combination of Southern (A, B), Western (C), and Northern (D) blot analysis. Genomic tail DNA from F1 offspring of the germline chimera was digested with HindIII and hybridized with genomic probe B, as depicted in Figure 1. Note that wt animals show a single band of 18 kb, whereas in heterozygous APLP1 mutants an additional band of the expected size of 14 kb was detected (A). Intercrossing of these heterozygous animals lead to homozygous mutants (B), as judged by analysis of DNA from mouse embryonic fibroblasts (MEF). The respective genotype is depicted above the blots. Western blots (C) of wt and APLP1−/− mice were prepared as described in Materials and Methods. Forty micrograms of brain extracts from two APLP1−/− mice (lanes 1, 2), a wt APLP1+/+littermate (lane 3), and a 129 Sv(ev) wt mouse (lane 4) were separated on a 12% standard Lämmli gel. Incubation with APLP1-specific antiserum CT11 followed by anti-rabbit horseradish peroxidase-linked secondary antibody showed a set of APLP-1-specific bands of 85–100 kDa for wt brain homogenates. In contrast, brain homogenates from APLP1−/− animals showed neither APLP1-specific bands of wt size nor any shorter polypeptides. The blot was developed with chemiluminescence reagents (ECL; Amersham). Marker proteins of the indicated sizes were from Bio-Rad (broad range rainbow markers). Total RNA was isolated from brain of either mutant or wt animals, and poly(A+) RNA was prepared from 120 μg of total RNA. Northern blot analysis (D) with an antisense RNA probe (cDNA position 1788–2360) lying downstream of the targeted deletion revealed a band of 2.4 kb on brain of wt animals, whereas no transcript was detected in organs from APLP1−/− mutant animals. After autoradiography, filters were stripped and rehybridized with a GAPDH probe to monitor loading (bottom panel).