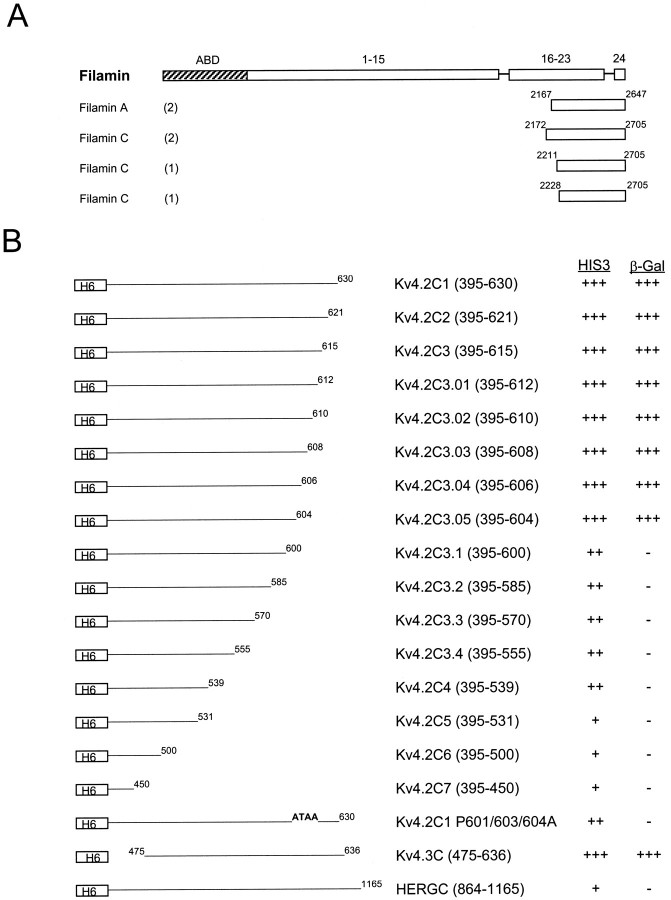
Fig. 1.
The domain structure of filamin and interaction with Kv4.2. A, Human cDNA clones isolated with a yeast two-hybrid screen, using the Kv4.2 C-terminal region (aa 395–630) as bait, are shown aligned below a schematic representation of the filamin domain structure. ABD, Actin binding domain;1–15, 16–23, and 24represent ∼96 aa repeats, each separated by hinge regions. Partial cDNAs from filaminA and filaminC genes were isolated. The numbers in parenthesesindicate the number of times each clone was isolated with the yeast two-hybrid screen. B, Sequence requirements in the Kv4.2 C-terminal region for interaction with filamin. FilaminC(aa 2172–2705), binding to Kv4.2C1 (aa 395–630), and deletion derivatives were assayed by HIS3/β-gal induction in the yeast two-hybrid system. Residues 601–604 are required for interaction with filamin; deletion and/or mutation of this region abolishes the interaction. Kv4.3, which contains the identical binding region, also interacts with filamin. The HERG C-terminal region (aa 864–1165) does bind filamin. The various bait fragments were tested for filamin binding by semiquantitative yeast two-hybrid interaction assays that were based on the degree of induction by the reporter genesHIS3 and β-gal. HIS3 activity was measured by the percentage of colonies growing on histidine-lacking medium as compared with the full-length Kv4.2 bait (Kv4.2C1): +++, >75%; ++, >50%; +, >25%. β-Gal activity was determined from the time that was taken for the colonies to turn blue in X-gal filter lift assays performed at room temperature: +++, <2 hr; ++, <3 hr; +, <4 hr; −, no significant activity.H6, Sixth transmembrane domain.