Figure 2.
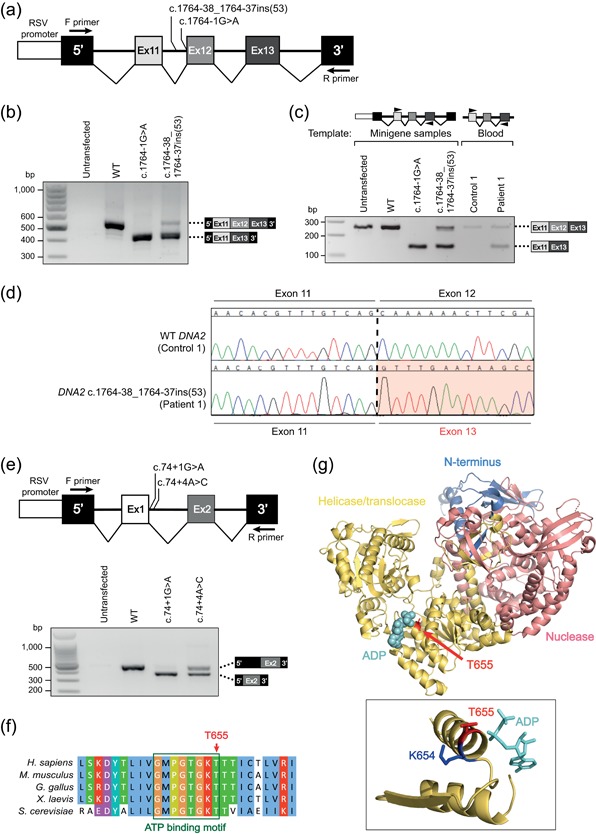
Transcriptional and structural consequences of DNA2 variants identified in MPD patients. (a) RHCglo minigene reporter constructs used in the minigene splicing assay to assess the effect of the DNA2 c.1764‐38_1764‐37ins(53) variant. A positive control for splicing disruption was generated by introducing a point mutation, abolishing the acceptor splice site of DNA2 intron 11. Arrows indicate primers used for RT‐PCR analysis. (b) c.1764‐38_1764‐37ins(53) affects splicing of DNA2 transcript. HeLa cells were transfected with minigene constructs, followed by RNA extraction, cDNA generation, and RT‐PCR analysis to assess DNA2 splicing patterns. RT‐PCR products amplified from the HeLa cDNA of the minigene assay samples using F and R primers (see panel a) were separated by agarose gel electrophoresis. Sanger sequencing analysis of cloned PCR products revealed that the product with higher electrophoretic mobility represents transcript lacking exon 12, and the lower mobility product the correctly spliced transcript. (c) The splicing defect caused by DNA2 c.1764‐38_1764‐37ins(53) is also detected in patient (P1) PBLs. PCR was performed using primers located in exon 11 and exon 13 of DNA2 (indicated by short black arrows) products using cDNA generated from minigene assay samples and from RNA extracted from P1 peripheral blood. Agarose gel electrophoresis and Sanger sequencing analysis of PCR products recapitulated the previous findings. (d) Sanger electropherograms demonstrate that the c.1764‐38_1764‐37ins(53) variant results in skipping of DNA2 exon 12. (e) Top: RHCglo minigene reporter constructs used in the minigene splicing assay to assess the effect of the DNA2 c.74+4A>C variant. As a control, the c.74+1G>A point mutation was introduced, abolishing the acceptor splice site of DNA2 intron 1. Bottom: DNA2 c.74+4A>C variant affects splicing of DNA2 transcript. PCR products amplified from HeLa cDNA of the minigene assay samples were separated by agarose gel electrophoresis. Sanger sequencing analysis of cloned PCR products revealed that the product with higher electrophoretic mobility represents transcript lacking exon 1 and the lower mobility product the correctly spliced transcript. (f) DNA2 threonine 655, mutated in MPD patient P3, is a highly conserved residue in the ATP binding motif (boxed in green) within the helicase/translocase domain. Alignment generated and visualized using Jalview multiple sequence alignment software (Waterhouse, Procter, Martin, Clamp, & Barton, 2009). (g) DNA2 p.K654 and p.T655 are important for ATP/ADP binding. Top: DNA2 protein domains (N‐terminus, nuclease and helicase/translocase) are represented in different colors (PDB ID: 5EAW; [Zhou et al., 2015]). ADP is shown in cyan spheres; p.T655, shown in red, indicates mouse DNA2 p.T656 (the equivalent of human p.T655). Below: this threonine residue contacts ADP, similar to p.K654 (p.K655 in mouse, indicated in blue = p.K654 in human DNA2). Substitution of K654 abolishes DNA2 ATPase activity. ADP, adenosine diphosphate; ATP, adenosine triphosphate; cDNA, complementary DNA; MPD, microcephalic primordial dwarfism; PBL, peripheral blood leukocytes; RT‐PCR, reverse‐transcription polymerase chain reaction
