ABSTRACT
The endotoxin of Gram-negative bacteria threatens the intestinal health of livestock. Ethyl pyruvate (EP) has been shown to regulate intestinal immunity and protect against cell and tissue damage. In this study, it was first verified that EP could reduce the secretion of IL-8, TNF-α, IL-6 and IL-1β in LPS-induced IPEC-J2 cells. Then, we used RNA sequencing (RNA-seq) to analyze the differentially expressed genes (DEGs) of inflammatory factors induced by LPS in IPEC-J2 cells. It was found that LPS induced the upregulation of 377 genes and the downregulation of 477 genes compared to Vehicle; LPS+EP induced the upregulation of 258 genes and the downregulation of 240 genes compared to Vehicle; and LPS+EP induced the upregulation of 373 genes and the downregulation of 188 genes compared to LPS (fold change > 1.5 and FDR < 0.01). Their enrichment pathways included the MAPK signaling pathway, PI3K-Akt signaling pathway, Toll-like receptor signaling pathway, and other pathways. Furthermore, the mRNA level of cytokines associated with inflammation and apoptosis enriched in the MAPK pathway was verified by qRT-PCR. Western blots and immunofluorescence revealed that EP significantly inhibited phosphorylated p38 and phosphorylated-ERK1/2 protein expression levels (P < 0.05). The apoptosis due to LPS reduced by EP was significantly inhibited, as shown by Annexin V-FITC/PI staining. According to the results, EP inhibited the expression of IL-8, TNF-α, IL-6 and IL-1β as well as apoptosis by inhibiting the phosphorylation of p38 and ERK1/2 in LPS-induced IPEC-J2 cells.
KEYWORDS: Ethyl pyruvate, RNA-seq, Porcine jejunal epithelial cells, Apoptosis, MAPK
Introduction
The intestinal diseases of animals have always affected the development of animal husbandry. LPS is lipopolysaccharide carried on the cell wall of Gram-negative bacteria, which is detrimental to the intestinal health of animals. LPS is ubiquitous and is released during bacterial growth and stationary phases and when bacterial death causes disintegration [1]. Many studies have shown that LPS induces intestinal inflammation, destroys intestinal structure and causes the release of inflammatory factors such as IL-6, IL-8, IL-1β, TNF-α and COX-2 [2–5]. Moreover, LPS activates the innate immune system and induces apoptosis [6]. A recent study shows that LPS promotes cell apoptosis through p38 and JNK pathways [7]. Thus, MAPK signaling pathway plays an important role in LPS-induced inflammatory responses and apoptosis [8–10]. It is important to find effective therapeutic targets for LPS-induced inflammatory bowel disease by studying the mechanism of infection.
Ethyl pyruvate (EP) has proven to be an effective compound for the treatment of intestinal inflammation infections. It is derived from pyruvate, an important metabolite in the tricarboxylic acid cycle. EP repairs tissue or cell damage and enhances immunity by anti-oxidation and reducing the secretion and expression of inflammatory factors induced by LPS such as IL-1β, IL-6 and IL-8 [11–13]. EP also has a therapeutic effect of repairing intestinal barrier on enteritis induced by external stimuli [14]. It has been reported that EP upregulated HO-1 through p38MAPK signaling pathway in the treatment of sepsis. This also works through Nrf2 transposition into the nucleus, following the combination of Nrf2 with the antioxidant response element of HO-1. This combination decreases LPS-induced iNOS by inhibiting the binding of p65 and p300 [15,16]. EP reduces the neuro-inflammatory factor matrix metalloproteinase-9 by inhibition of on the phosphorylation of p38 MAPK, ERK, and Akt [17]. EP also inhibited the increase of LPS induced high mobility group box 1(HMGB1) by up regulating SIRT1 and inhibiting the phosphorylation of STAT pathway [18]. In addition, TLR4 is a receptor specifically binding to LPS in TLRs [19]. Secretion of pro-inflammatory cytokines stimulated by LPS are decreased through inhibition of TLR4 and phosphorylated p65 by in SW480 and THP-1 cells [20]. Another anti-inflammatory method of EP is increasing the anti-inflammatory cytokines IL-10 meanwhile down-regulation of pro-inflammatory factors [21]. Additionally, studies have shown that EP inhibits apoptosis induced by myocardial ischemia and reduces white matter injury induced by LPS in rats by anti-apoptosis mechanisms [22,23]. Thus, EP inhibits inflammation and cell apoptosis through multiple pathways.
However, the anti-inflammatory mechanism of EP acting on intestinal epithelial cells has not been studied in detail. In this study, we examined the effects of EP on LPS-induced inflammatory cytokines in IPEC-J2 cells and analyzed the mechanism of EP regulating inflammation through RNA sequencing. According to our RNA-seq findings, MAPK is closely related to IPEC-J2 cell inflammation. We used qRT-PCR to further verify the expression trends of the differential genes and the phosphorylation levels of the major proteins p38 and ERK of the MAPK pathway through western blots and immunofluorescence. The protective effect of EP on LPS-induced apoptosis was observed by fluorescence microscopy and flow cytometry. This study aimed at investigating the functions and pathways by which EP reduces the expression of inflammatory factors in LPS-induced IPEC-J2 cells.
Materials and methods
Reagents
IPEC-J2 cells were cultured in DMEM/F12 medium (Gibco, USA) containing 10% fetal bovine serum (Hyclone, USA) and 1% penicillin/streptomycin (100 U/mL and 100 mg/mL, respectively) (Beyotime). Ethyl pyruvate was purchased from Sigma-Aldrich [natural, ≥ 98%, FG (Aldrich)]. Anti-phospho-p38 MAPK rabbit mAb, anti-p38 MAPK rabbit mAb, anti-phospho-ERK1/2 rabbit mAb and anti-ERK1/2 rabbit mAb were purchased from Cell Signaling Technology (USA); the secondary antibody HRP-labeled goat anti-rabbit IgG (H + L) was from Beyotime (China); and the ERK inhibitor U0126 and the p38 MAPK kinase inhibitor SB203580 were purchased from Sigma (USA).
Cell culture
The cells were divided into three groups: control group, LPS group and LPS+EP group, with three replicates in each group. Each sample of 1 × 106 cells was incubated for 12 h at 37 °C with 5% CO2, and then, the control was treated with medium without serum. The LPS group was treated with 10 μg/mL LPS in medium without serum for 12 h and the LPS+EP group was co-treated with 10 μg/mL LPS and 2.5 mM EP in medium without serum for 12 h. All the RNA samples were collected.
ELISA assay
The levels of the pro-inflammatory factors IL-8, TNF-α, IL-6 and IL-1β in the cell culture supernatant samples were assayed with a commercial enzyme-linked immunosorbent assay kit (JingMei Biological Technology, China) according to the manufacturer’s instructions. Different concentrations of standards (50 μL/well) and 10 μL samples in 40 μL sample diluent were added into wells. Blank wells received 10 μL instead of samples. Then, we added 100 μL enzyme labeling reagent per well (except for the blank wells) and incubated the plate at 37 °C for 1 h. We then removed the liquid in the wells and washed the wells for 30 s with wash buffer, then discarded the buffer, repeated it 5 times, and then dried the wells. Chromogenic agent A was added (50 μL per well), and then, chromogenic agent B of the same volume was added to each well. We mixed the liquid lightly and incubated the plate at 37 °C for 15 min and then stopped the reaction with stop solution. We zeroed the ELISA microplate reader with the blank wells, and then, the absorbance values were determined at 450 nm. We drew the standard curve and calculated the concentration of samples.
RNA extraction and RNA-seq analysis
After cell incubation was complete, the cells were washed 3 times with cold PBS and then lysed with Trizol. After total RNA was extracted, eukaryotic mRNA was enriched by Oligo (dT) beads. Then, the enriched mRNA was fragmented into short fragments using fragmentation buffer and reverse transcribed into cDNA with random primers. Second-strand cDNA was synthesized by DNA polymerase I, Rnase H, dNTP and buffer. Then, the cDNA fragments were purified with a QiaQuick PCR extraction kit, end repaired, poly(A) added, and ligated to Illumina sequencing adapters. The ligation products were size-selected by agarose gel electrophoresis, PCR amplified, and sequenced using Illumina HiSeqTM 4000.
Analysis of differentially expressed genes (DEGs)
For bioinformatics analysis, the original reading containing the adapter or low quality (Qvalue ≤ 10) was removed and mapped to the pig reference genome (assembly SSCROFA 11–1). The EdGER package (http://www-rord.org/) was used to identify differentially expressed genes between different samples or groups. We identified genes with a fold change ≥ 1.5 and a false discovery rate p ≤ 0.05 compared with significant DEGs. Then, we analyzed the enrichment of DEGs using GO functions and the KEGG pathway database.
qRT-PCR
For verification of differential expression genes, the total RNA of each well was treated with 0.2 mL Trizol reagent and was reverse transcribed into cDNA with a PrimeScriptTM RT reagent Kit (RR037A, Takara Bio Inc., Shiga, Japan). Then, the mRNA expression was measured as the reference by using TB green Premix Ex Taq as the fluorescent dye (RR420A, Takara Bio Inc., Shiga, Japan). The PCR cycling conditions were 95 °C (30 s) for 1 cycle, 95 °C (5 s), and 58 °C (34 s) for 40 cycles. Relative quantifications of the mRNA expression of the inflammatory factor genes are shown as the comparative threshold cycle number for each sample (2–ΔΔCT). Gene expression was compared with the corresponding β-actin level. All the primer pairs used for qRT-PCR were designed and synthesized from Sangon (China), and sequences are shown in Table 1.
Table 1.
Primers of real-time PCR.
| Gene | Primer (5ʹ-3ʹ) | Accession |
|---|---|---|
| BDNF | F:GGCATAGACAAGAGGCACTGGAAC R:TCCTTATGAACCGCCAGCCAATTC |
NM_214259.2 |
| JUND | F:GCTCGAACGTCTCATCATCCAGTC R:TCGGCGAACTCCTGCTCCTC |
XM_021083521.1 |
| MKK6 | F:CGTCAAGCCTTCCAACGTCCTG R:CGACAAGGTAGCCGCTGATTCC |
GQ169690.1 |
| MST1 | F:TTGGCAAGAGGATGTGGCAGATG R:GCAGCCGTGTATGAGGTGAGTG |
XM_003132215.4 |
| DUSP8 | F:CTGTTGGACTGTCGCTCCTTCTTC R:CGCCGCACGATGGTACTGAAG |
XM_021082598.1 |
| TBK1 | F:ACTGGAAAGCCTTCTGGTGC R:ACAGGCATGTCTCCACTCCA |
NM_001105292.1 |
| β-Actin | F:CAGGTCATCACCATCGGCAACG R:GACAGCACCGTGTTGGCGTAGAGGT |
U07786 |
Western blotting analysis
To determine the level of p38 protein phosphorylation, cell protein samples were collected with 0.1 mL RIPA with 10 μL PMSF (Beyotime catalog no. P0013B). The lysate was centrifuged at 10, 000 g for 30 min at 4 °C, and then, the supernatant was transferred into a new tube. All samples were mixed with 4× loading buffer and boiled for 10 min. For detecting phosphorylated p38 and ERK1/2, the protein to be tested was separated on a 12% SDS-polyacrylamide gel for 20 min at 80 V and then 1 h at 120 V. Next, we transferred the total protein from the gel to the PVDF membrane at 75 V for 90 min, and then, PVDF membranes were blocked by 5% fat-free milk powder at 37 °C for 2 hours. The PVDF membranes were incubated with primary antibody (dilution, 1:1000) at 4 °C overnight. Then, they were washed in TBST for 10 min three times. Next, membranes were incubated with the corresponding HRP-linked secondary antibodies (dilution, 1:1000) for 1 h at 37 °C and washed with TBST as above. The protein bands were observed by a super-enhanced chemiluminescence (ECL) Plus detection system (P0018, Beyotime, China) and observed with X-ray film (Clinx Science Instruments Co., Ltd., Shanghai, China).
Immunofluorescence assay
IPEC-J2 cells (5 × 104 cells/well in 96-wells plates) were cultured in 96-wells plates with different treatments to detect the location of the p-p38 and p-ERK. After the treatment, the cells were washed with PBS, following which they were fixed with 80% acetone for 30 min at 4 °C and washed with PBS for 5 min. The fixed cells were then permeabilized in 0.5% Triton X-100 for 10 min at room temperature, washed with PBS for 5 min, and then treated with 10% goat serum in PBS for 60 min. The permeabilized cells were then treated with Phospho-p38 MAPK (Thr180/Tyr182) (D3F9) XP® Rabbit mAb (1:100) (Cell Signaling Technology) or Phospho-p44/42 MAPK (ERK1/2) (Thr202/Tyr204) (D13.14.4E) XP® Rabbit mAb (Cell Signaling Technology) for 60 min at room temperature and washed with PBS for 5 min. The cells were then incubated in a 1:100 dilution of Alexa Fluor 568-labeled goat anti-rabbit antibody (Beyotime, USA) for 60 min at room temperature and washed in PBS for 5 min. Cells were stained with DAPI staining solution (C1005, Beyotime, USA) for 10 min at 37 °C and then washed with PBS. Finally, the cell samples were taken photos by EVOS FL Auto.
Detection of apoptosis
IPEC-J2 cells (5 × 104 cells/well in a 96-wells plate) were seeded in 6-well plates. We incubated the cells overnight, treated different wells of cells separately, and continued to incubate for 24 h. We discarded the medium, washed the cells with PBS, and then incubated with an Annexin V-FITC/PI kit for 10 min. Finally, we observed apoptosis fluorescence by fluorescence microscopy.
Data analysis
All data were obtained from at least three independent experiments and expressed as the means ± standard deviations (SD). The differences and statistical significance of the gene expression ratios of drug-treated versus normal samples were analyzed using an unpaired Student’s t-test or GLM by (SPSS.20). A P-value less than 0.01–0.02 was considered statistically significant. Different letters indicate significant difference at P < 0.05.
Results
Effects of EP on secretion of inflammatory factor in LPS induced IPEC-J2 cells
The pro-inflammatory cytokines IL-8, TNF-α, IL-6 and IL-1β not only play an important role in LPS-induced inflammation but are also markers of inflammation. The concentration of IL-8, TNF-α, IL-6 and IL-1 in cell supernatant was detected by ELISA. As Figure 1 shows, LPS induced significant increases in the secretion of IL-8, TNF-α, IL-6 and IL-1β, and EP reduced these secretions significantly.
Figure 1.
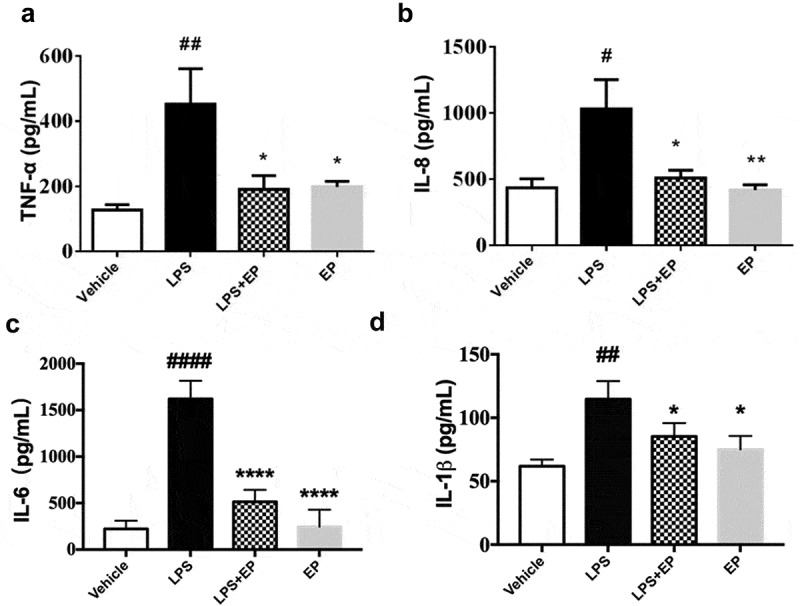
Effects of EP on secretion of inflammatory factors in LPS induced IPEC-J2 cells. (a) Concentration of TNF-α in IPEC-J2 cells; (b) Concentration of IL-8 in IPEC-J2 cells; (c) Concentration of IL-6 in IPEC-J2 cells; (d) Concentration of IL-1β in IPEC-J2 cells. IPEC-J2 cells were treated by 10 μg/mL LPS, 10 μg/mL LPS+2.5 mM EP or 2.5 mM EP respectively for 12 h. #p means significant compared with control, *p means significant between LPS group. Values are mean ± SD.
GO biological function analysis of DEGs
According the GO terms shown in Figure 2, differential expression genes (DEGs) were divided into three groups: those involved in biological processes, those that are cellular components, and those with molecular functions. We found that most of the DEGs were enriched on binding and cellular processes. According to GO enrichment analysis, we discovered that 34 genes were enriched on immune system process.
Figure 2.
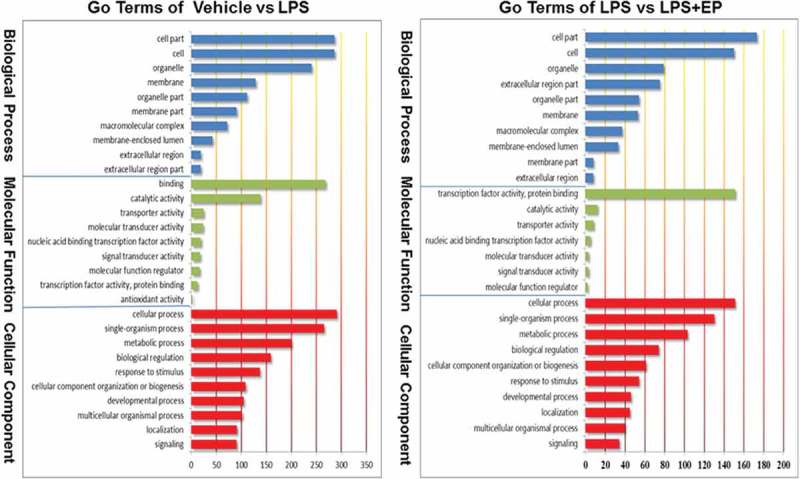
(Continued).
Figure 2.
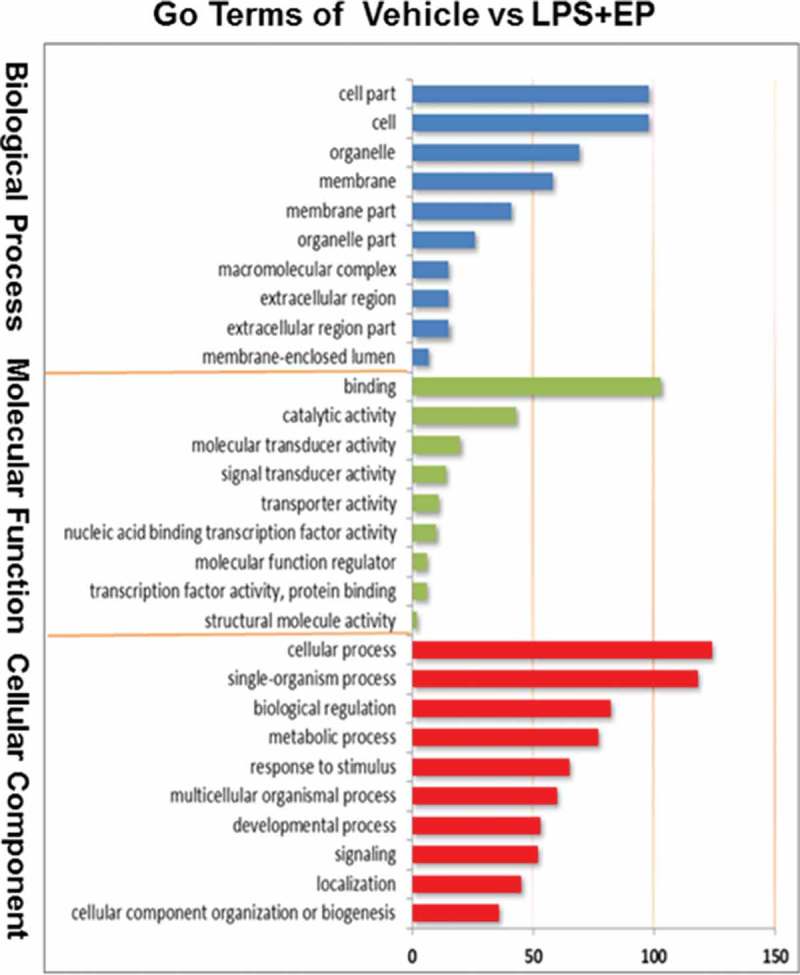
Go Terms of biological function analysis among Vehicle, LPS and LPS+EP groups.
KEGG pathway analysis of DEGs
We performed numerical analysis of DEGs as shown in Figure 3. It was found that LPS induced the upregulation of 377 genes and the downregulation of 477 genes compared to Vehicle; LPS+EP induced the upregulation of 258 genes and the downregulation of 240 genes compared to Vehicle; and LPS+EP induced the upregulation of 373 genes and the downregulation of 188 genes compared to LPS (fold change > 1.5 and FDR < 0.01). To study the EP regulatory pathways, we performed a KEGG enrichment pathway analysis (Figure 4(a,b)). We collected immune-related pathways: MAPK signaling pathway, PI3K-Akt signaling pathway, Toll-like receptor signaling pathway, cytokine-cytokine receptor interaction, JAK-STAT signaling pathway, inflammatory mediator regulation of TRP channels, NF-kappa B signaling pathway, TNF signaling pathway, NOD-like receptor signaling pathway, and mTOR signaling pathway. Among all the inflammatory pathways, the MAPK signaling pathway was enriched with the largest number of differential genes. This finding suggested that the MAPK signaling pathway may be the key pathway to regulate LPS-induced inflammatory signaling of IPEC-J2 cells. All the differential genes enriched on inflammatory pathways are shown in Table 2.
Figure 3.
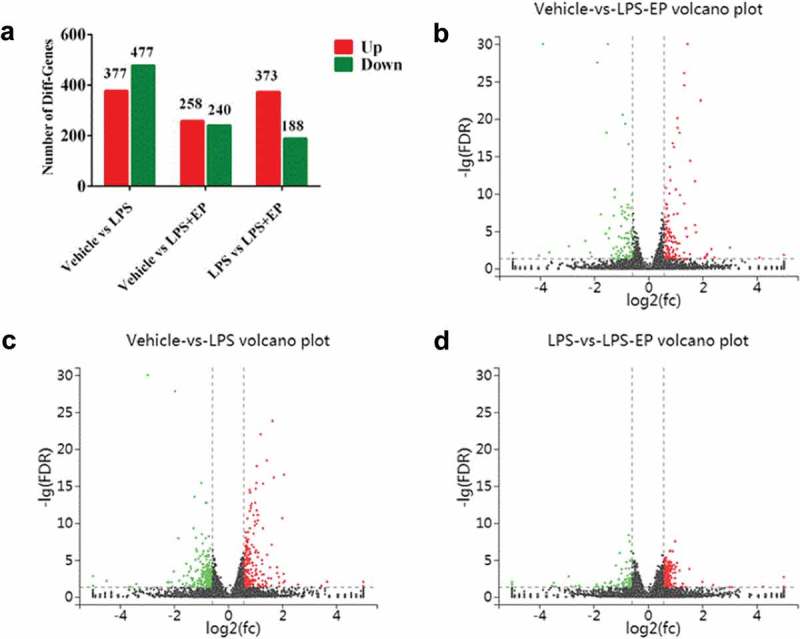
Up-regulation genes and down-regulation genes. (a)Numbers of DEGs between groups compared each other. (b-d) Vocano plot of DEGs. The x-axis indicates the difference in expression level on a log2 (fold change). The y-axis represents corresponding false discovery rate on a negative Lg (FDR). Red represents up-regulation gene and green means down-regulation genes.
Figure 4.
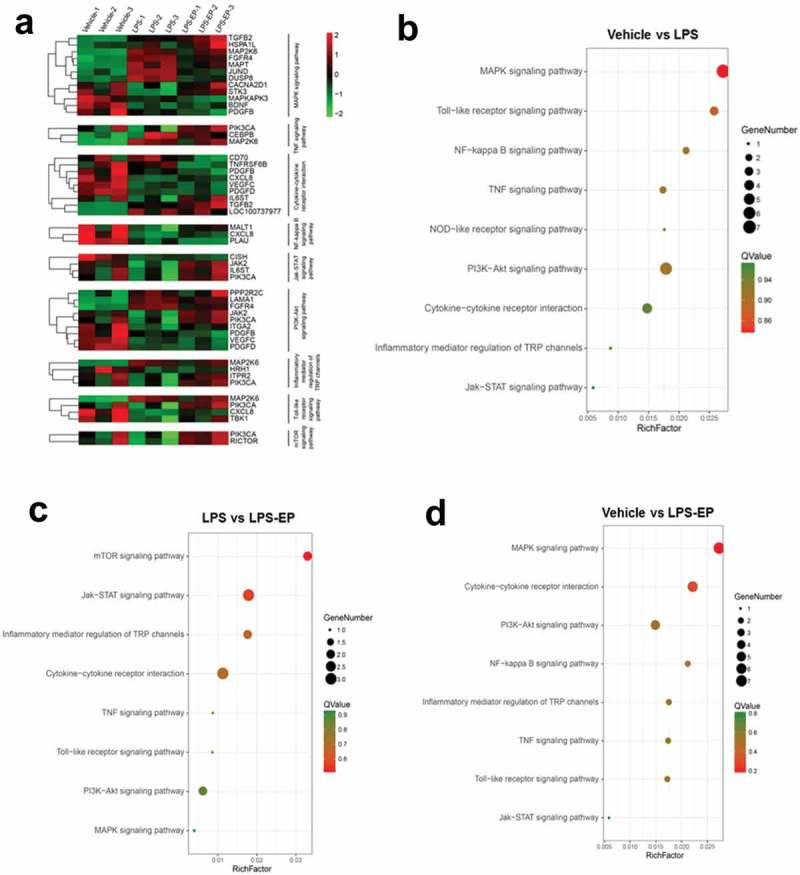
(a) Heatmaps of all the differential genes in KEGG inflammatory pathways. (b-d) Bubbles of KEGG pathways for differential gene enrichment. The circle presented gene number. The color of circles indicates the Q value.
Table 2.
Pathway enrichment analysis of the differential genes.
| KEGG Pathway | Pathway ID | Differential Gene (Vehicle v.s LPS) | Differential Gene (Vehicle v.s LPS+EP) | Differential Gene (LPS v.s LPS+EP) |
|---|---|---|---|---|
| MAPK signaling pathway | ko04010 | BDNF, MST1/2, DUSP, FGFR4, MKK6, JUND, MAPT | BNDF, PDGF, FGFR4, HSP72, MAPKAPK2, MKK6, TGFB | CACNA2D1 |
| PI3K-Akt signaling pathway | ko04151 | PPP2R2, FGFR4, VEGFC_D, PDGFC_D, LAMA1_2, ITGA2 | FGFR4, VEGFC_D, PDGFC_D, LAMA1_2, PDGFB | PIK3C, JAK2 |
| Toll-like receptor signaling pathway | ko04620 | MKK6, TBK1, IL8 | MKK6, IL8 | PIK3C |
| Cytokine-cytokine receptor interaction | ko04060 | TNFRSF10, VEGFC_D, PDGFC_D, IL8 | TNFRSF6B, VEGFC_D, PDGFC_D, IL8, TGFB2, PDGFB | TNFRSF6B, TNFSF7, IL6ST |
| Jak-STAT signaling pathway | ko04630 | CISH | CISH | PIK3C, JAK2, IL6ST |
| Inflammatory mediator regulation of TRP channels | ko04750 | MKK6 | HRH1, MKK6 | PIK3C, ITPR2 |
| NF-kappa B signaling pathway | ko04064 | MALT1, IL8 | IL8, uPA | |
| TNF signaling pathway | ko04668 | MKK6, CEBPB | MKK6, CEBPB | PIK3C |
| NOD-like receptor signaling pathway | ko10030 | IL8 | IL8 | |
| mTOR signaling pathway | ko04150 | PIK3C, RICTOR |
Validation of DEGs data by qRT-PCR
To verify the DEGs identified by sequencing, we randomly selected 6 genes from the significant DEGs. The 6 upregulated genes were as follows: MST1, BDNF, JUND, TBK1, MKK6, and DUSP-8. Data are expressed as fold change in gene expression. Figure 5 shows that gene expression levels agree in terms of the sequencing and RT-qPCR data, which confirmed that the sequencing data are reliable.
Figure 5.
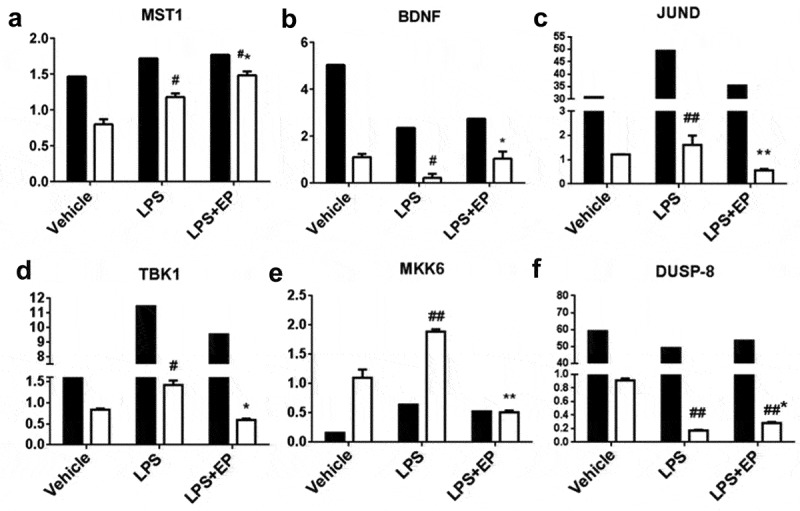
Validation of the RNA-Seq expression profiles of genes randomly selected from DEGG by RT-qPCR. Black bars represent FPKM, white bars represent fold change. FPKM (Fragments Per Kilobase Million) normalized values as gene expression in RNA-seq. #p means significant compared with control, *p means significant between LPS group. Values are mean ± SD.
EP reduced inhibited inflammatory factors phosphorylation by inhibiting p38 and ERK1/2
To detect the inhibitory effect of EP on the activation of the MAPK pathway, we measured the phosphorylated protein levels of p38 and ERK1/2 by western blotting and immunofluorescence. As shown in Figures 6 and 7, EP effectively inhibited LPS-induced phosphorylation of p38 and ERK1/2. The p38 and ERK1/2 phosphorylated protein in the LPS+EP group were significantly lower in the LPS group as seen on western blot. In Figure 7, phosphorylated p38 and ERK1/2 were observed in the LPS group. However, there was no detectable fluorescence in the Vehicle, LPS+EP or EP groups.
Figure 6.
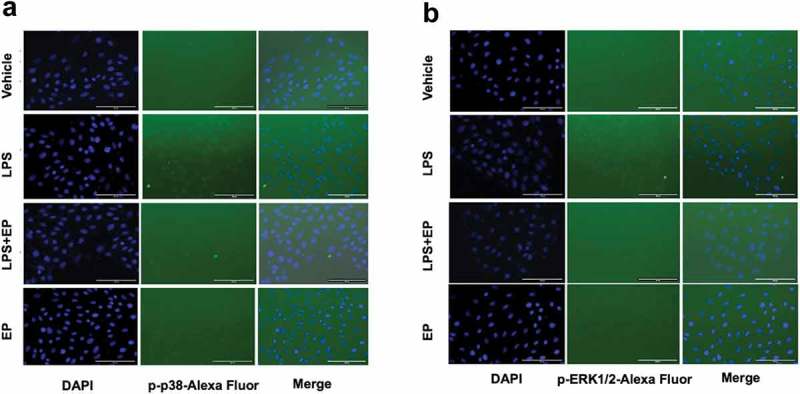
(a-b) Transient expression of the p-p38 and p-ERK1/2 protein in IPEC-J2 cells by fluorescence microscopy. IPEC-J2 cells were incubated overnight and treated by 10 μg/mL LPS, 10 μg/mL LPS+2.5 mM EP or 2.5 mM EP respectively for 12 h. P-p38 and p-ERK1/2 protein fluorescence was determined by fluorescence microscope.
Figure 7.
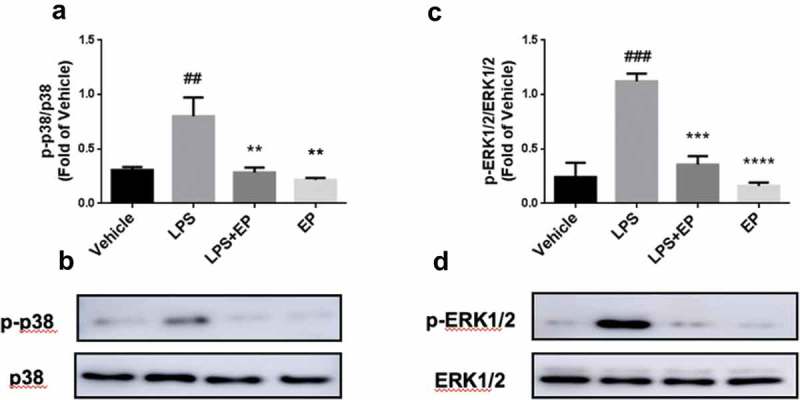
Phosphoprotein levels of p38 and ERK1/2. (a-b) Phosphoprotein levels of p38 in IPEC-J2 cells. (c-d) Phosphoprotein levels of ERK1/2 in IPEC-J2 cells. Cells were treated by 10 μg/mL LPS, 10 μg/mL LPS+2.5 mM EP or 2.5 mM EP respectively for 12 h. #p means significant compared with control, *p means significant between LPS group. Values are mean ± SD.
EP inhibits the expression of inflammatory factors through p38 and ERK1/2
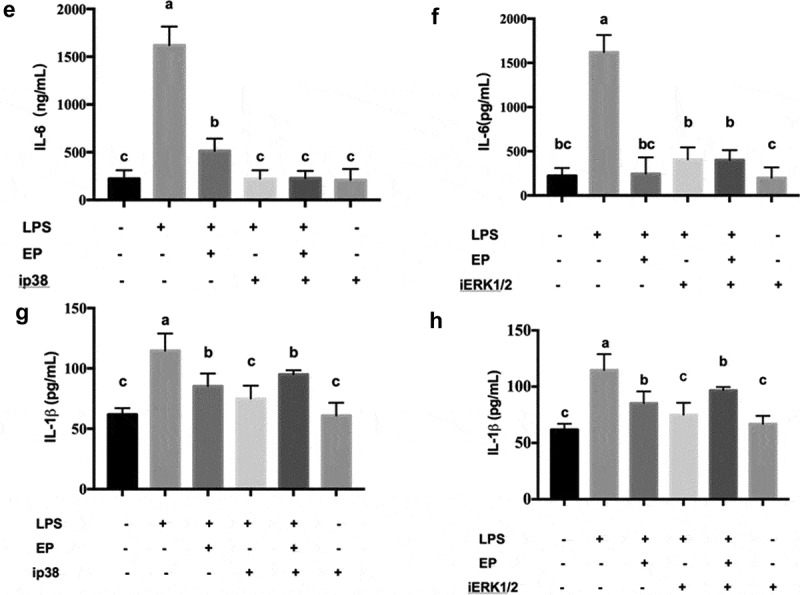
To further investigate the mechanism of EP’s inhibition of the expression of inflammatory factors by the p38 and ERK1/2 pathways, the concentrations of IL-8, TNF-α, IL-6 and IL-1β in IPEC-J2 were determined by ELISA. The results in Figure 8 show that IL-8, TNF-α, IL-6 and IL-1β decreased after treatment with the inhibitor of p38 and ERK1/2. This finding indicates that EP inhibited IL-8, TNF-α, IL-6 and IL-1β expression via the p38 and ERK1/2 MAPK pathways.
Figure 8.
(Continued).
Figure 8.
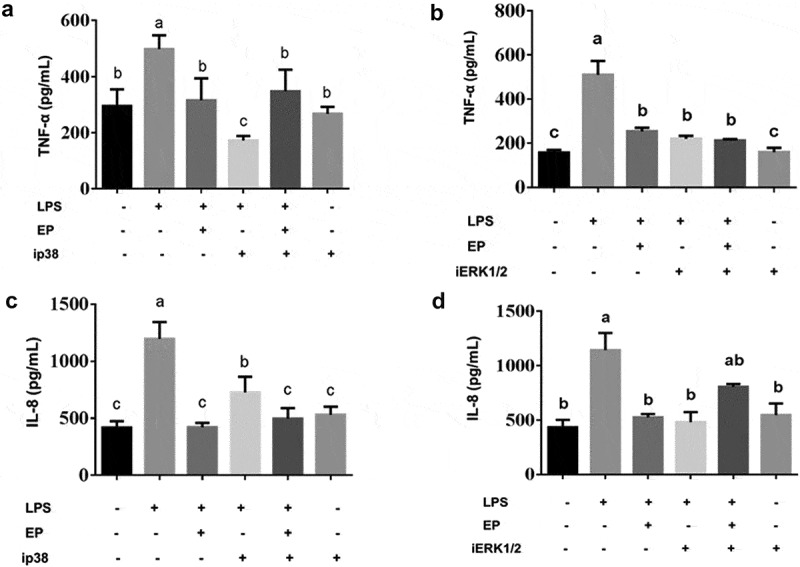
Effects of p38 and ERK1/2 inhibitors on the secretion of inflammatory factors by EP in LPS induced IPEC-J2 cells. ip38 represents p38 inhibition and iERK1/2 represents ERK1/2 inhibition. (a-b) Effects of p38 and ERK1/2 inhibition on secretion of TNF-α. (c-d) Effects of p38 and ERK1/2 inhibition on secretion of IL-8. (e-f) Effects of p38 and ERK1/2 inhibition on secretion of IL-6. (g-h) Effects of p38 and ERK1/2 inhibition on secretion of IL-1β. Cells were pre-incubated with 10 μM p38(SB203580) and 20 μM ERK1/2(U0126) inhibitors for 1 h and then treated by 10 μg/mL LPS or 10 μg/mL LPS+2.5 mM EP respectively for 12 h. Different values between groups are shown with capital letters (a, b, c; p < 0.05). Values are mean ± SD.
EP protects LPS-induced IPEC-J2 apoptosis
We also examined the effect of EP on LPS-induced apoptosis by flow cytometry and fluorescence staining. As shown in Figure 9, after LPS treatment both the nucleus and the cytoplasm showed obvious characteristics of apoptosis. There were no detected apoptotic cells in the control and EP groups. In the LPS+EP group, the number of apoptotic cells was significantly reduced compared with the LPS group.
Figure 9.
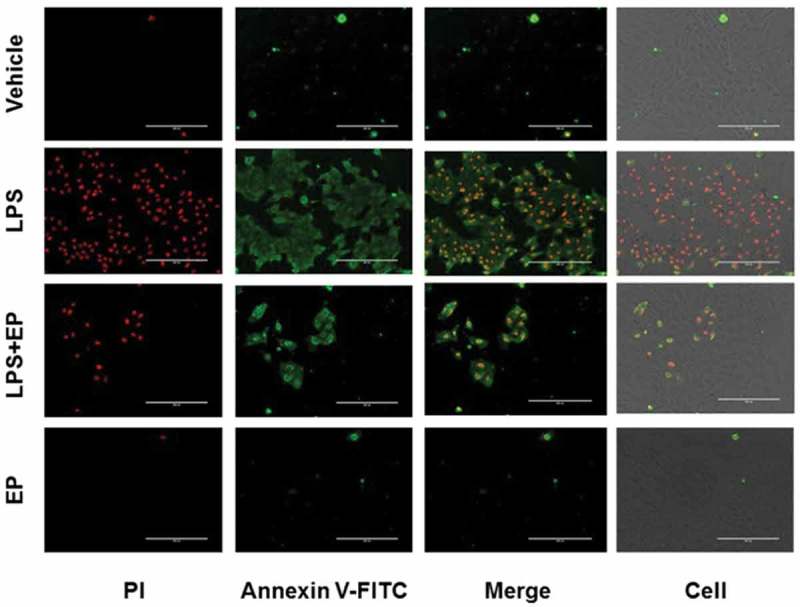
Effect of EP on LPS-induced apoptosis of IPEC-J2 cells. IPEC-J2 cells were incubated and treated by 10 μg/mL LPS, 10 μg/mL LPS+2.5 mM EP or 2.5 mM EP respectively for 12 h, then cells were incubated with Annexin V-FITC/PI kit for 10 min. Finally, observed apoptosis fluorescence by fluorescence microscope.
Discussion
LPS induces inflammatory factors and apoptosis in IPEC-J2 cells. EP is an anti-inflammatory and anti-apoptosis substance which repairs of cell and tissue damage that has been proved by many studies, but the mechanism of EP treatment on inflammatory intestinal epithelial cells is not clear [22,24,25]. In this study, RNA-seq was used to analyzed the differential expression gene enrichment pathways in which LPS stimulated IPEC-J2 cells and EP exerted anti-inflammatory effects.
TNF-α and interleukins are typical pro-inflammatory factors and rapidly secrete and produce inflammation when the body is stimulated by exogenous stimulation [26]. LPS induces oxidative stress and promotes the expression of pro-inflammatory factors such as TNF-α, IL-6 and IL-1β[27]. Because the secretion of TNF-α will further stimulate the production of other inflammatory factors, excessive secretion of inflammatory factors such as IL-1β, IL-2 and IL-8 induced intestinal mucosal injury when TNF-α has an overabundance production [28–30]. Accordingly, the inflammatory factors play typical roles in intestinal immunity. As Figure 1 shows, EP inhibited the secretion of the inflammatory cytokines IL-8, TNF-α, IL-6 and IL-1β induced by LPS in IPEC-J2 cells.
To analyze the mechanism of EP on LPS induced IPEC-J2 cell inflammation, we sequenced the mRNA with RNA-seq. The results showed all the DEGs, which were then subjected to GO biological function analysis and KEGG enrichment pathways. GO analysis demonstrated the pathways which were divided into three categories involving biological processes, cellular components and molecular functions. The most enriched pathways of DEGs are related to cell parts, transcription factor binding, and protein binding.
KEGG enrichment pathway analysis showed that DEGs and enriched major immunological pathways included the MAPK signaling pathway, the PI3K-Akt signaling pathway, and the Toll-like receptor signaling pathway. In the Vehicle vs LPS and Vehicle vs LPS+EP comparisons, MAPK was the main signaling pathway that had the most DEGs to regulate the inflammatory response. The main DEGs in the MAPK signaling pathway are shown in Table 2 and include BDNF, MST1/2, DUSP, MKK6 and JUND, as well as others. BDNF (brain-derived neurotrophic factor) and JUND are involved in protecting cells from apoptosis due to cytokines [31,32]. In our results, EP prevented the decrease of BDNF mRNA expression induced by LPS (Figure 5). MST1/2, DUSP and MKK6 are all cytokines that play an important role in innate immune regulation [33–35]. Previous research showed that MST1/2 can regulate the function of T cells, promote the transport of B cells and lack of MST1/2 will make B cell fail to circulate to lymph and bone marrow [36,37]. DUSP stands for dual-specificity phosphatases. DUSP1, DUSP4, and DUSP6 are involved in the formation of mammary stem cells and overexpression of DUSP6 in breast cancer cells [38]. DUSP2 also mediates the dephosphorylation of JNK, while DUSP4 and DUSP16 can promote the phosphorylation of ERK1/2 [39]. Similarly, the absence of DUSP8 will increase the activation of ERK1/2 [40,41]. Figure 5 shows that the LPS-induced decline of DUSP8 was significantly inhibited by EP. MKK6 is a regulatory kinase in the upstream of p38MAPK, which mainly promotes phosphorylation of p38 [42,43]. In our results, EP reduced MKK6 expression induced by LPS (Figure 5). TANK-binding kinase 1 (TBK1) mainly associated with autoimmune diseases, and a study has found that TBK1 is overexpressed in patients with lupus erythematosus [44]. TBK1 is also an upstream molecule of inducible NO synthase (iNOS), cyclooxygenase (COX-2), and TNF-α[45]. In addition, TBK1 is activated by viral infection and inflammatory mediators. Following infection, TBK1 participates in the regulation of type I interferon and NF-κB and MAPK signal transduction [46]. All the above DEGs demonstrated that the mechanism by which EP causes inhibition of inflammation may involve inhibition of p38 and ERK1/2 activation by preventing the phosphorylation of MAPKK family signal molecules such as MKK6 and by increasing the expression of DUSP (Figure 10).
Figure 10.
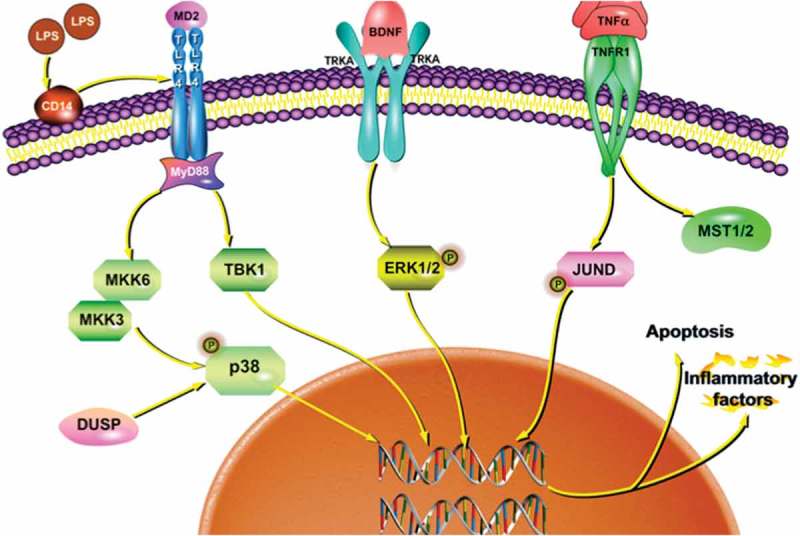
Signaling pathways of inflammation and apoptosis in LPS induced cells by EP regulation.
Furthermore, western blotting, immunofluorescence and MAPK signaling pathway inhibition tests were performed to confirm the role of the MAPK pathway in EP’s regulation of inflammation. In many reports, the MAPK pathway is one of the main pathways for LPS to induce inflammation [47,48]. LPS activates MyD88-mediated host immune responses after binding to TLR4 receptors when LPS invades the body [19]. Furthermore, activation of the MAPK pathway promotes the release of NO and pro-inflammatory cytokines such as IL-1β and TNF-α[49]. However, TNF-α and IL-1β also irritate the MAPK pathway to further promote the expression of IL-8 and IL-6 via activating p38 and ERK1/2 [50–52]. Thus, inhibition of phosphorylation of p38 and ERK1/2 can effectively inhibit the MAPK pathway in order to avoid pathological changes caused by excessive release of inflammatory factors. In general, inflammatory pathways are selected as targets for anti-inflammatory drugs [53]. According to our results, EP significantly inhibited the phosphorylation of p38 and ERK1/2 in LPS-induced IPEC-J2 cells (Figures 6 and 7). Moreover, inhibition of p38 and ERK1/2 pathways revealed reduced expression of IL-8, TNF-α, IL-6 and IL-1β. In conclusion, EP plays an anti-inflammatory role by inhibiting p38 and ERK1/2.
Apoptosis plays a key role in ensuring the healthy survival of multicellular organisms and an important role in the normal development of individuals. It is important in the tissue differentiation, organ development and homeostasis of multicellular organisms [54,55]. When cells are during inflammation, survival of cells depends on the process of apoptosis [56]. On the basis of a previous study, LPS induces apoptosis of macrophages by inducing the secretion of TNF-α[57]. In addition, activation or inactivation of the MAPK pathway regulates the cell apoptosis process [58]. In this study, the protective effect of EP on LPS-induced apoptosis of IPEC-J2 cells was examined by fluorescence microscopy (Figure 9). These results indicated that EP prevented cell apoptosis by inhibiting ERK1/2 phosphorylation.
Conclusion
In conclusion, we demonstrated that EP inhibited LPS-induced inflammatory cytokines in IPEC-J2 cells. Furthermore, we used RNA-seq to compare differential gene expression and enriched pathways of control, LPS and LPS+EP treatment groups. RNA-seq results showed that EP inhibited LPS-induced secretion of inflammatory factors in IPEC-J2 cells induced by inhibiting p38 and ERK1/2 of the MAPK pathway via preventing MKK6 and DUSP activation. We proved that EP protected IPEC-J2 cells from apoptosis. Verification of these pathways provides new ideas for the treatment of intestinal infections.
Funding Statement
This work was supported by the Heilongjiang Provincial Science and Technology Department [QC2015018];National Natural Science Foundation of China [31472104];National Natural Science Foundation of China [31501914];National Natural Science Foundation of China [31672434];Postdoctoral Research Foundation of China [2015M571385].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Wells JE, Russell JB.. The effect of growth and starvation on the lysis of the ruminal cellulolytic bacterium Fibrobacter succinogenes. Appl Environ Microbiol. 1996;62:1342–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cao S, Zhang Q, Wang CC, et al. LPS challenge increased intestinal permeability, disrupted mitochondrial function and triggered mitophagy of piglets. Innate Immun. 2018;24:221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wu W, Wang S, Liu Q, et al. Cathelicidin-WA attenuates LPS-induced inflammation and redox imbalance through activation of AMPK signaling. Free Radic Biol Med. 2018;129:338–353. [DOI] [PubMed] [Google Scholar]
- [4].Gao Z, Liu X, Wang W, et al. Characteristic anti-inflammatory and antioxidative effects of enzymatic- and acidic- hydrolysed mycelium polysaccharides by Oudemansiella radicata on LPS-induced lung injury. Carbohydr Polym. 2019;204:142–151. [DOI] [PubMed] [Google Scholar]
- [5].Macrophages L, Ekstrelerinin F, Etkileri UMA.. Anti-inflammatory effects of pelargonium endlicherianum Fenzl. Extracts in. Turk J Pharm Sci. 2018;15:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lu Z, Li Y, Ru JH, et al. Interaction of palmitate and LPS regulates cytokine expression and apoptosis through sphingolipids in human retinal microvascular endothelial cells. Exp Eye Res. 2018;178:61–71. [DOI] [PubMed] [Google Scholar]
- [7].Xiao Z, Liu L, Tao W, et al. Clostridium tyrobutyricum protect intestinal barrier function from LPS-induced apoptosis via P38/JNK signaling pathway in IPEC-J2 cells. Cell Physiol Biochem. 2018;46:1779–1792. [DOI] [PubMed] [Google Scholar]
- [8].Sung J, Jeon H, Kim IH, et al. Anti-inflammatory effects of stearidonic acid mediated by suppression of NF-κB and MAP-kinase pathways in macrophages. Lipids. 2017;52:781–787. [DOI] [PubMed] [Google Scholar]
- [9].Guo C, Yang RJ, Jang K, et al. Protective effects of pretreatment with quercetin against lipopolysaccharide-Induced apoptosis and the inhibition of osteoblast differentiation via the MAPK and Wnt/β-Catenin pathways in MC3T3-E1 Cells. Cell Physiol Biochem. 2017;43:1547–1561. [DOI] [PubMed] [Google Scholar]
- [10].Liu H, Zhou K, Liao L, et al. Lipoxin A4 receptor agonist BML-111 induces autophagy in alveolar macrophages and protects from acute lung injury by activating MAPK signaling. Respir Res. 2018;19:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shen M, Lu J, Dai W, et al. Corrigendum to “Ethyl pyruvate ameliorates hepatic ischemia-reperfusion injury by inhibiting intrinsic pathway of apoptosis and autophagy.”. Mediators Inflamm. 2016;2016:1–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ozacmak HS, Ozacmak VH, Turan I. Ethyl pyruvate prevents from chronic cerebral hypoperfusion via preserving cognitive function and decreasing oxidative stress, caspase 3 activation and IL-1β level. Bratislava Med J. 2018;119:469–475. [DOI] [PubMed] [Google Scholar]
- [13].Zhao J, Cheng J, Li C, et al. Ethyl pyruvate attenuates CaCl2 -induced tubular epithelial cell injury by inhibiting autophagy and inflammatory responses. Kidney Blood Press Res. 2018;43(5):1585–1595. [DOI] [PubMed] [Google Scholar]
- [14].Sappington PL, Han X, Yang R, et al. Ethyl pyruvate ameliorates intestinal epithelial barrier dysfunction in endotoxemic mice and immunostimulated caco-2 enterocytic monolayers. J Pharmacol Exp Ther. 2003;304:464–476. [DOI] [PubMed] [Google Scholar]
- [15].Jang HJ, Kim YM, Tsoyi K, et al. Ethyl pyruvate induces heme oxygenase-1 through p38 mitogen-activated protein kinase activation by depletion of glutathione in RAW 264.7 Cells and improves survival in septic animals. Antioxid Redox Signal. 2012;17:878–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim SW, Lee HK, Shin JH, et al. Up-down regulation of HO-1 and iNOS gene expressions by ethyl pyruvate via recruiting p300 to Nrf2 and depriving It from p65. Free Radic Biol Med. 2013;65:468–476. [DOI] [PubMed] [Google Scholar]
- [17].Lee EJ, Kim HS. Inhibitory mechanism of MMP-9 gene expression by ethyl pyruvate in lipopolysaccharide-stimulated BV2 microglial cells. Neurosci Lett. 2011;493:38–43. [DOI] [PubMed] [Google Scholar]
- [18].Kim YM, Park EJ, Kim JH, et al. Ethyl pyruvate inhibits the acetylation and release of HMGB1 via effects on SIRT1/STAT signaling in LPS-activated RAW264.7 cells and peritoneal macrophages. Int Immunopharmacol. 2016;41:98–105. [DOI] [PubMed] [Google Scholar]
- [19].Shang L, Wang T, Tong D, et al. Prolyl hydroxylases positively regulated LPS-induced inflammation in human gingival fibroblasts via TLR4/MyD88-mediated AKT/NF-κB and MAPK pathways. Cell Prolif. 2018;51(6):e12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang FC, Pei JX, Zhu J, et al. Overexpression of HMGB1 A-box reduced lipopolysaccharide-induced intestinal inflammation via HMGB1/TLR4 signaling in vitro. World J Gastroenterol. 2015;21:7764–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Yu D-H, Noh D-H, Song R-H, et al. Ethyl pyruvate downregulates tumor necrosis factor alpha and interleukin (IL)-6 and upregulates IL-10 in lipopolysaccharide-stimulated canine peripheral blood mononuclear cells. J Vet Med Sci. 2010;72:1379–1381. [DOI] [PubMed] [Google Scholar]
- [22].Guo J, Zhang J, Luo X, et al. Effects of ethyl pyruvate on cardiac function recovery and apoptosis reduction after global cold ischemia and reperfusion. Exp Ther Med. 2014;7:1197–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang Y, Yin P, Huang S, et al. Ethyl pyruvate protects against lipopolysaccharide-induced white matter injury in the developing rat brain. Int J Dev Neurosci. 2013;31:181–188. [DOI] [PubMed] [Google Scholar]
- [24].Dong W, Cai B, Peña G, et al. Ethyl pyruvate prevents inflammatory responses and organ damage during resuscitation in porcine hemorrhage. Shock. 2010;34:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang R, Zhu S, Tonnessen TI. Ethyl pyruvate is a novel anti-inflammatory agent to treat multiple inflammatory organ injuries. J Inflamm (United Kingdom). 2016;13:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wang Q, Zhang B, Yu JL. Farrerol inhibits IL-6 and IL-8 production in LPS-stimulated human gingival fibroblasts by suppressing PI3K/AKT/NF-κB signaling pathway. Arch Oral Biol. 2016;62:28–32. [DOI] [PubMed] [Google Scholar]
- [27].Li M-Y, Sun L, Niu X-T, et al. Astaxanthin protects lipopolysaccharide-induced inflammatory response in Channa argus through inhibiting NF-κB and MAPKs signaling pathways. Fish Shellfish Immunol. 2018;86:280–286. [DOI] [PubMed] [Google Scholar]
- [28].Indaram AVK, Visvalingam V, Locke M, et al. Mucosal cytokine production in radiation-induced proctosigmoiditis compared with inflammatory bowel disease. Am J Gastroenterol. 2000;95:1221–1225. [DOI] [PubMed] [Google Scholar]
- [29].Sugimoto M, Yamaoka Y, Furuta T. Influence of interleukin polymorphisms on development of gastric cancer and peptic ulcer. World J Gastroenterol. 2010;16:1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ishiguro Y. Mucosal proinflammatory cytokine production correlates with endoscopic activity of ulcerative colitis. J Gastroenterol. 1999;34:66–74. [DOI] [PubMed] [Google Scholar]
- [31].Xia D, Li W, Zhang L, et al. RNA interference-mediated knockdown of brain-derived neurotrophic factor (BDNF) promotes cell cycle arrest and apoptosis in B-cell lymphoma cells. Neoplasma. 2014;61:523–532. [DOI] [PubMed] [Google Scholar]
- [32].Wang T, Li P, Ma X, et al. MicroRNA-494 inhibition protects nucleus pulposus cells from TNF-α-induced apoptosis by targeting JunD. Biochimie. 2015;115:1–7. [DOI] [PubMed] [Google Scholar]
- [33].Juknat A, Pietr M, Kozela E, et al. Microarray and pathway analysis reveal distinct mechanisms underlying cannabinoid-mediated modulation of LPS-induced activation of BV-2 microglial cells. PLoS One. 2013;8(4):e61462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Du X, Shi H, Li J, et al. Mst1/Mst2 regulate development and function of regulatory T cells through modulation of Foxo1/Foxo3 stability in autoimmune disease. J Immunol. 2014;192:1525–1535. [DOI] [PubMed] [Google Scholar]
- [35].Liu J, Han L, Li B, et al. F-box only protein 31 (FBXO31) negatively regulates p38 mitogen-activated protein kinase (MAPK) signaling by mediating lysine 48-linked ubiquitination and degradation of Mitogen-activated protein kinase kinase 6 (MKK6). J Biol Chem. 2014;289:21508–21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Li J, Du X, Shi H, et al. Mammalian Sterile 20-like Kinase 1 (Mst1) enhances the stability of forkhead Box P3 (Foxp3) and the function of regulatory T cells by modulating Foxp3 acetylation. J Biol Chem. 2015;290:30762–30770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Alsufyani F, Mattoo H, Zhou D, et al. The Mst1 Kinase Is required for Follicular B Cell Homing and B-1 B Cell Development. Front Immunol. 2018;9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Boulding T, Wu F, McCuaig R, et al. Differential roles for DUSP family members in epithelial-to-mesenchymal transition and cancer stem cell regulation in breast cancer. PLoS One. 2016;11(2):e0148065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xu T, Wu X, Chen Q, et al. The anti-apoptotic and cardioprotective effects of salvianolic acid A on rat cardiomyocytes following ischemia/reperfusion by DUSP-mediated regulation of the ERK1/2/JNK pathway. PLoS One. 2014;9:e102292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Linte CA, Camp JJ, Rettmann ME, et al. DUSP8 regulates cardiac ventricular remodeling by altering ERK1/2 signaling. Circulation Research. 2015;119:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ferguson BS, Nam H, Morrison RF. Dual-speci fi city phosphatases regulate mitogen-activated protein kinase signaling in adipocytes in response to in fl ammatory stress. Cell Signal. 2019;53:234–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lin S, Liu K, Zhang Y, et al. Pharmacological targeting of p38 MAP-Kinase 6 (MAP2K6) inhibits the growth of esophageal adenocarcinoma. Cell Signal. 2018;51:222–232. [DOI] [PubMed] [Google Scholar]
- [43].Liu Z, Shi S, Zhu H, et al. Novel ASK1 Inhibitor AGI-1067 attenuates AGE-induced fibrotic response by suppressing the MKKs/p38 MAPK pathway in human coronary arterial smooth muscle cells. Int Heart J. 2018;59(6):1416–1424. [DOI] [PubMed] [Google Scholar]
- [44].White EK, Wakeland Q-Z, Li N, et al. Cutting edge: inhibiting TBK1 by cutting edge: Inhibiting TBK1 by compound II ameliorates autoimmune disease in mice. J Immunol Mater Suppl J Immunol KU Leuven Univ Libr. 2015;195(10):4573–4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yang Y, Lee GJ, Yoon DH, et al. ERK1- and TBK1-targeted anti-inflammatory activity of an ethanol extract of Dryopteris crassirhizoma. J Ethnopharmacol. 2013;145:499–508. [DOI] [PubMed] [Google Scholar]
- [46].Möser CV, Stephan H, Altenrath K, et al. TANK-binding kinase 1 (TBK1) modulates inflammatory hyperalgesia by regulating MAP kinases and NF-κB dependent genes. J Neuroinflammation. 2015;12:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Park J-Y, Park S-D, Koh YJ, et al. Aqueous extract of Dipsacus asperoides suppresses lipopolysaccharide-stimulated inflammatory responses by inhibiting the ERK1/2 signaling pathway in RAW 264.7 macrophages. J Ethnopharmacol. 2018;231:253–261. [DOI] [PubMed] [Google Scholar]
- [48].Han JM, Lee EK, Gong SY, et al. Sparassis crispa exerts anti-inflammatory activity via suppression of TLR-mediated NF-κB and MAPK signaling pathways in LPS-induced RAW264.7 macrophage cells. J Ethnopharmacol. 2018;231:10–18. [DOI] [PubMed] [Google Scholar]
- [49].Cho KH, Kim DC, Yoon CS, et al. Anti-neuroinflammatory effects of citreohybridonol involving TLR4-MyD88-mediated inhibition of NF-kB and MAPK signaling pathways in lipopolysaccharide-stimulated BV2 cells. Neurochem Int. 2016;95:55–62. [DOI] [PubMed] [Google Scholar]
- [50].Liu X, Ye F, Xiong H, et al. IL-1β upregulates IL-8 production in human Müller cells through activation of the p38 MAPK and ERK1/2 signaling pathways. Inflammation. 2014;37:1486–1495. [DOI] [PubMed] [Google Scholar]
- [51].Jijon HB, Buret A, Hirota CL, et al. The EGF receptor and HER2 participate in TNF-α-dependent MAPK activation and IL-8 secretion in intestinal epithelial cells. Mediators Inflamm. 2012;2012:207398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gezer U, Cetinkaya M, Isin M, et al. Lipopolysaccharide increases IL-6 secretion via activation of the ERK1/2 signaling pathway to up-regulate RANKL gene expression in MLO-Y4 cells. Cell Biol Int. 2014;41(1):84–92. [DOI] [PubMed] [Google Scholar]
- [53].Zhuang Y, Liu J, Ma P, et al. Tamarixinin a alleviates joint destruction of rheumatoid arthritis by blockade of MAPK and NF-κB activation. Front Pharmacol. 2017;8:538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hassan M, Watari H, AbuAlmaaty A, et al. Apoptosis and molecular targeting therapy in cancer. Biomed Res Int. 2014;2014:150845. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [55].Zaman S, Wang R, Gandhi V. Targeting the apoptosis pathway in hematologic malignancies. Leuk Lymphoma. 2014;55:1980–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Messer JS. The cellular autophagy/apoptosis checkpoint during inflammation. Cell Mol Life Sci. 2017;74:1281–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xaus J, Comalada M, Valledor AF, et al. LPS induces apoptosis in macrophages mostly through the autocrine production of TNF-alpha. Blood. 2000;95:3823–3831. [PubMed] [Google Scholar]
- [58].Feng Y, Fang L, Du Z, et al. Wip1 regulates SKOV3 cell apoptosis through the p38 MAPK signaling pathway. Mol Med Rep. 2017;15:3651–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]


