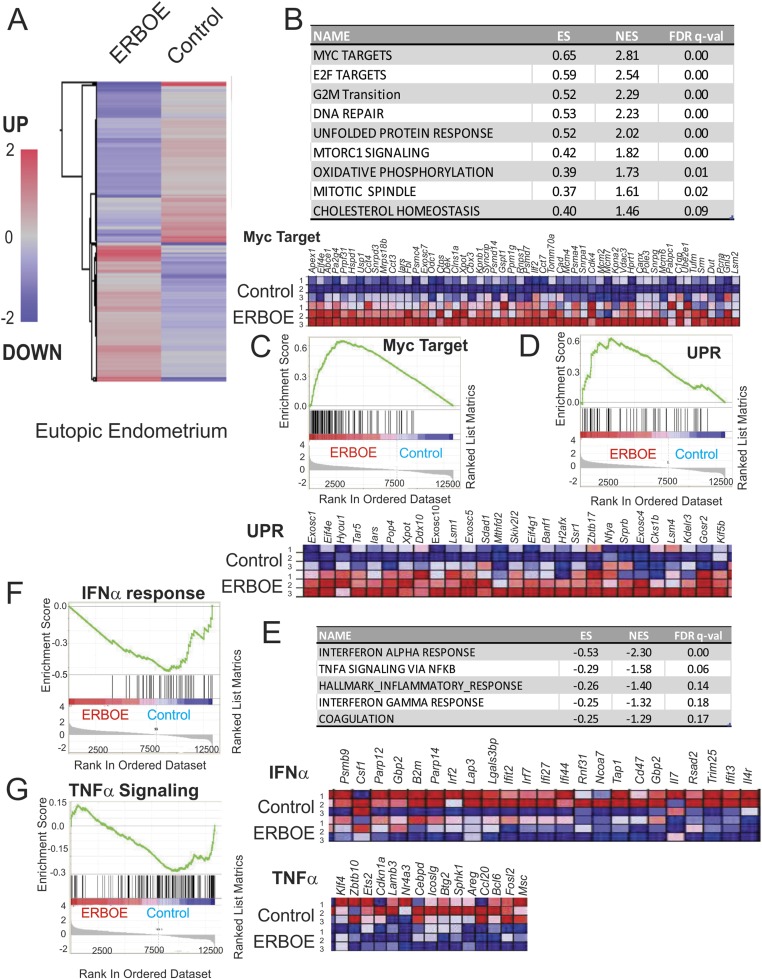
Figure 3.
Overexpression of ERβ changes cellular pathways in the eutopic endometrium to cause endometrial dysfunction. (A) Heat map of differential gene expression profiles (>1.5-fold upregulation or downregulation) in the ERβ-overexpressing eutopic endometrium vs control eutopic endometrium (average gene expression levels from nine samples per group). (B) Cellular pathways >1.5-fold upregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium determined by GSEA. (C) List of Myc target genes elevated in the ERβ-overexpressing eutopic endometrium compared with the control eutopic endometrium. Relative expressions of the Myc target gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (D) Genes involved in UPR signaling that are upregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium. Relative expressions of the UPR signaling gene set in control vs ERβ-overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (E) Cellular pathways >1.5-fold downregulated in the ERβ-overexpressing eutopic endometrium compared with the eutopic control endometrium determined by GSEA. (F) GSEA analysis revealed that ERβ overexpression reduced gene signatures associated with the IFNα signaling in ectopic lesions. Relative expressions of the IFNα signaling gene set in control vs ERβ-overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. (G) GSEA analysis revealed that ERβ overexpression reduced gene signatures associated with TNFα/NF-κB signaling in ectopic lesions. Relative expressions of the TNFα/NF-κB signaling gene set in control vs ERβ overexpressing ectopic lesions (N = 3) are shown in order of their signal/noise ratio rank. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; UPR, unfolded protein response.