Figure 6. Non-linear amplification of sequence-dependent pausing by HRDC.
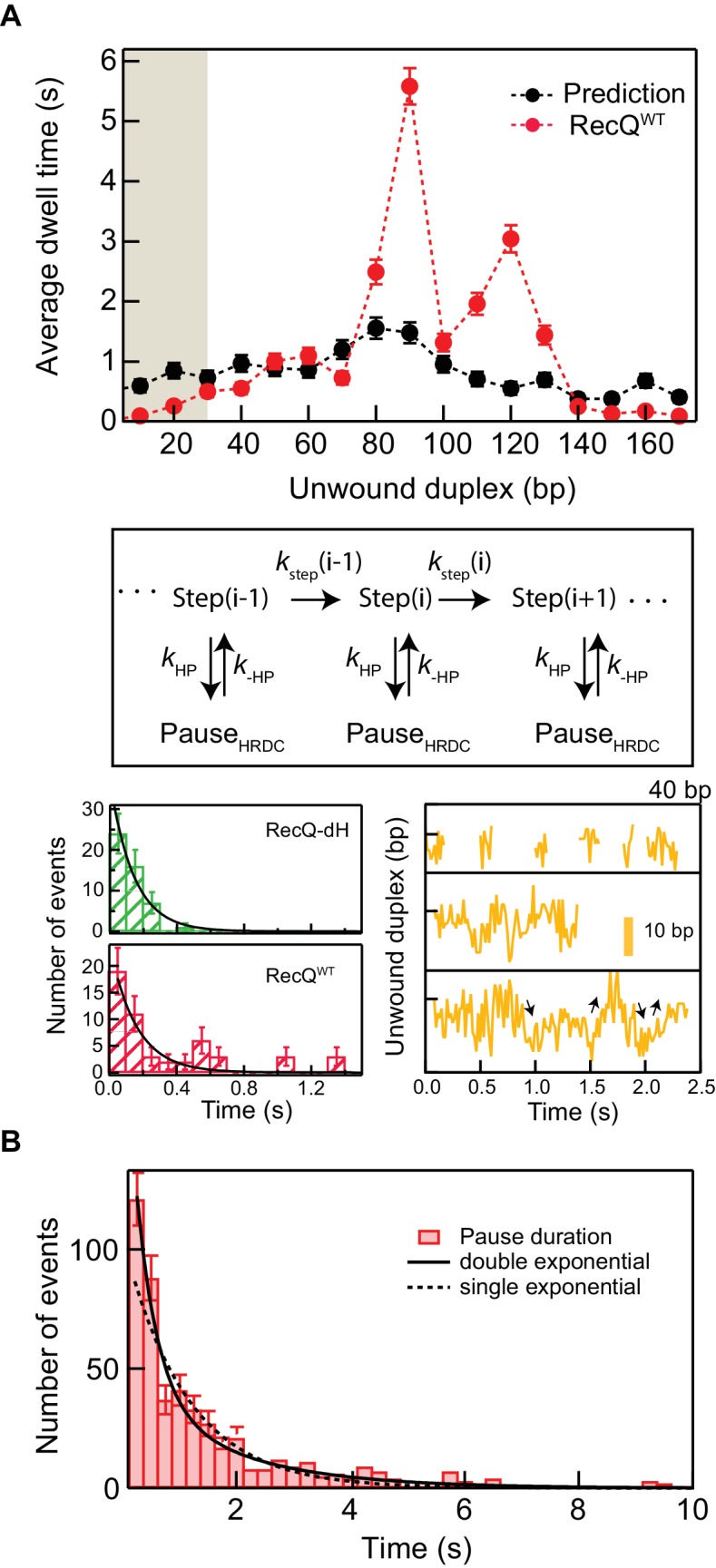
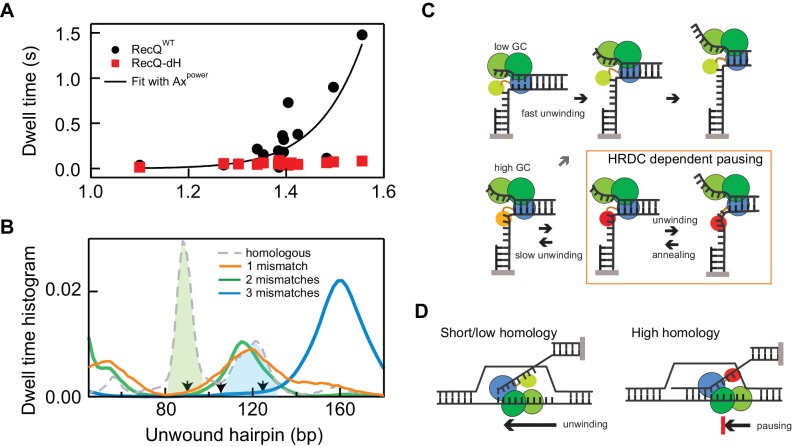
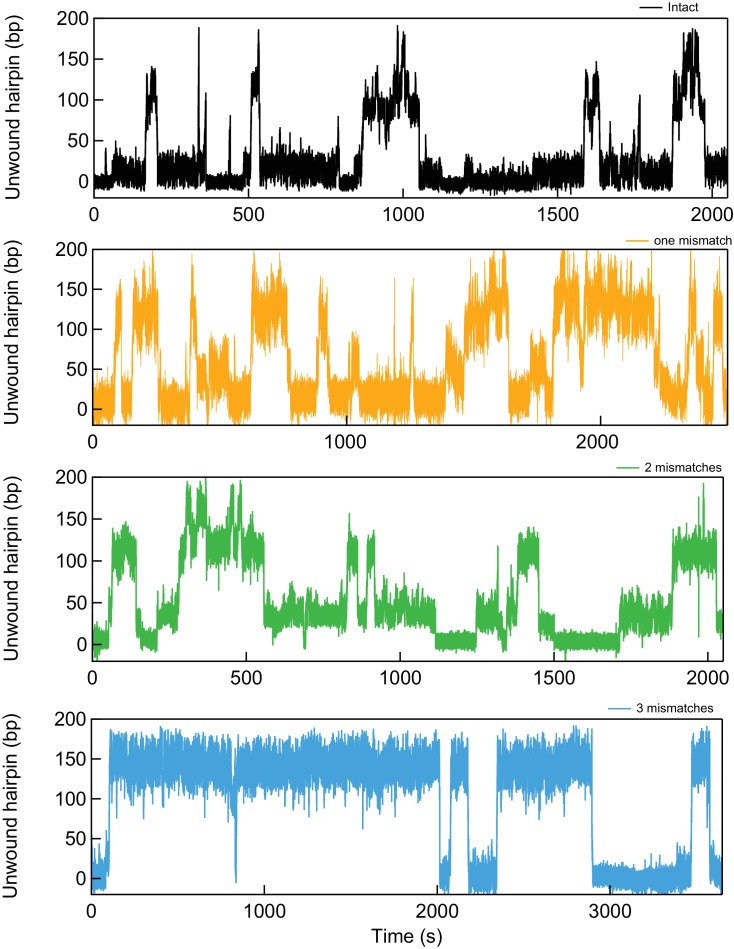
(A) The average dwell times of RecQWT and RecQ-dH plotted as a function of the exponential of the average base-pair energy over a 10 bp window. The non-linearity of RecQWT dwell time data was analyzed with a power law function (y = A·xP solid line). The fit returned A = 7 ± 0.9 (x10−4) and Power = 17.2 ± 3.2 (Errors indicate standard deviations of fitting parameters). (B) Dwell-time histograms of RecQ unwinding the 174 bp DNA hairpin with zero mismatches, that is perfect homology, one mismatch (90 bp), two mismatches (90 and 104 bp), and three mismatches (90, 104, and 124 bp). The prominent peak around 90 bp (green shaded region) shown in the dwell time histogram of the intact DNA unwinding by RecQWT was significantly reduced in the hairpin with a single mismatch and the additional peak around 120 bp (blue shaded region) was further suppressed by the third mismatch at 124 bp. The mismatch sites are indicated as arrows. (C) Model for D-loop homology discrimination via HRDC-mediated non-linear amplification of sequence-dependent pausing by RecQ. At regions of low GC content (upper row), RecQ rapidly unwinds duplex DNA and the HRDC remains in a weak binding ssDNA mode (light green HRDC). At regions of high GC content (lower row), RecQ pauses and the HRDC can switch to a strong ssDNA binding mode (orange HRDC). The subsequent binding of the HRDC to the displaced ssDNA (red HRDC) results in stabilization of the GC-induced pauses. As ssDNA is under tension or otherwise constrained, this interaction effectively hinders the movement of the RecQ core, resulting in short-range (5–10 bp) repetitive unwinding and annealing of DNA. (D) HRDC-dependent pausing regulates D-loop disruption in a homology dependent manner. RecQ can quickly unwind an invading strand of short or only partially homologous DNA, whereas HRDC-dependent pausing slows down unwinding and prevents disruption of an invading strand with an extended homology.
Figure 6—figure supplement 1. HRDC-dependent pausing kinetics are inconsistent with a simple kinetic competition mechanism of pausing.