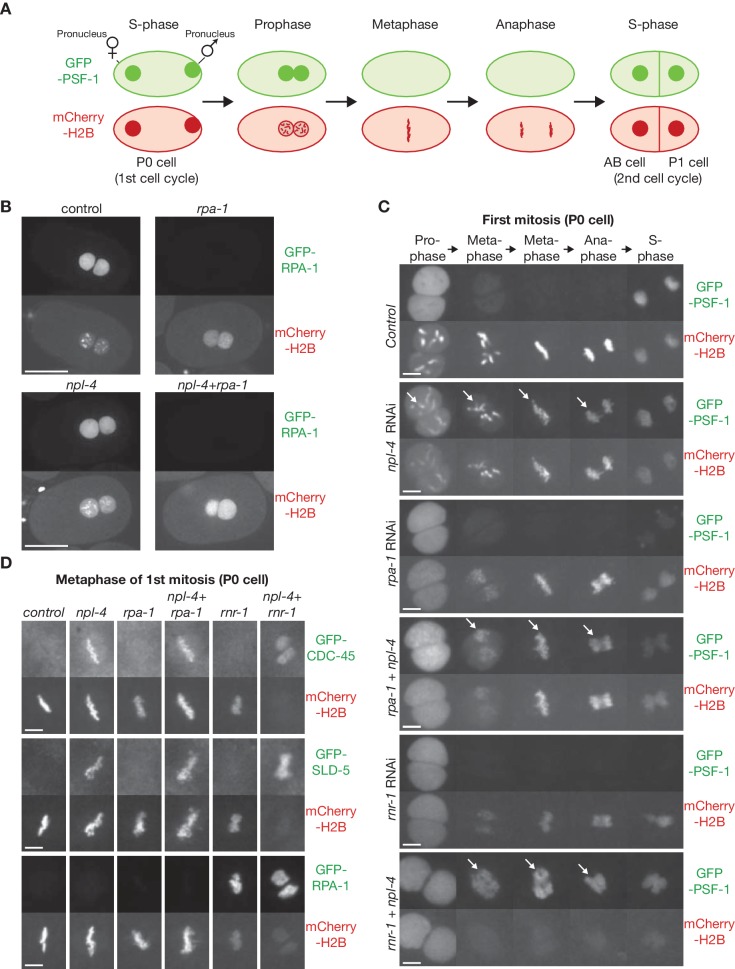
Figure 1. Incomplete DNA replication in C. elegans early embryos leads to unloading of the CMG helicase from chromatin by the CDC-48_UFD-1_NPL-4 segregase.
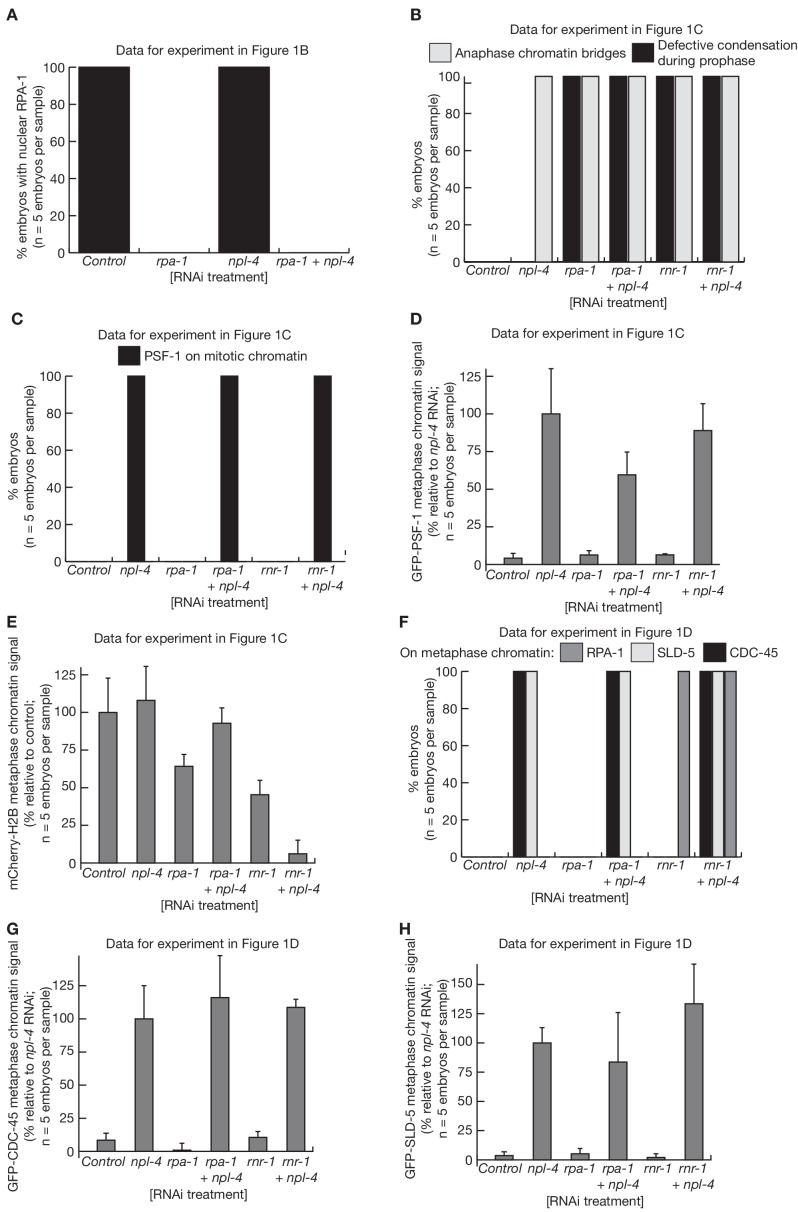
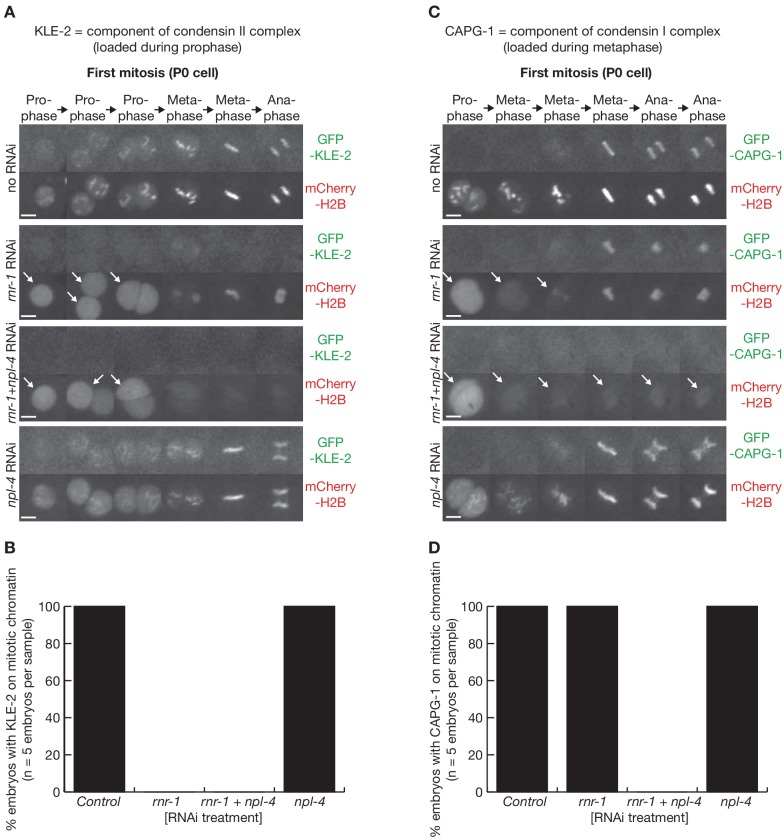
(A) Illustration of the first cell cycle in the C. elegans early embryo. (B) Worms expressing GFP-RPA-1 and mCherry-Histone H2B were exposed to the indicated RNAi treatments (control = no RNAi). Images of whole embryos are shown, during prophase of the first embryonic cell cycle (two minutes before nuclear envelope breakdown), in order to illustrate the efficiency of RPA-1 depletion. (C) Worms expressing GFP-PSF-1 and mCherry-Histone H2B were exposed to the indicated RNAi treatments, before time-lapse imaging of the first mitosis in isolated embryos. The arrows indicate persistence of CMG on mitotic chromatin. (D) Worms expressing the indicated GFP-tagged replication proteins, together with mCherry-Histone H2B, were exposed to the same range of RNAi treatments as in (C). Examples are shown of the metaphase stage of the first embryonic mitosis. Scale bars = 20 µm in (B) and 5 µm in (C and D). Quantification of microscopy data is provided in Figure 1—figure supplement 1.