Fig. 4. Gain of HNF4A binding during hepatoblast to hepatocyte differentiation accompanies histone and DNA modifications.
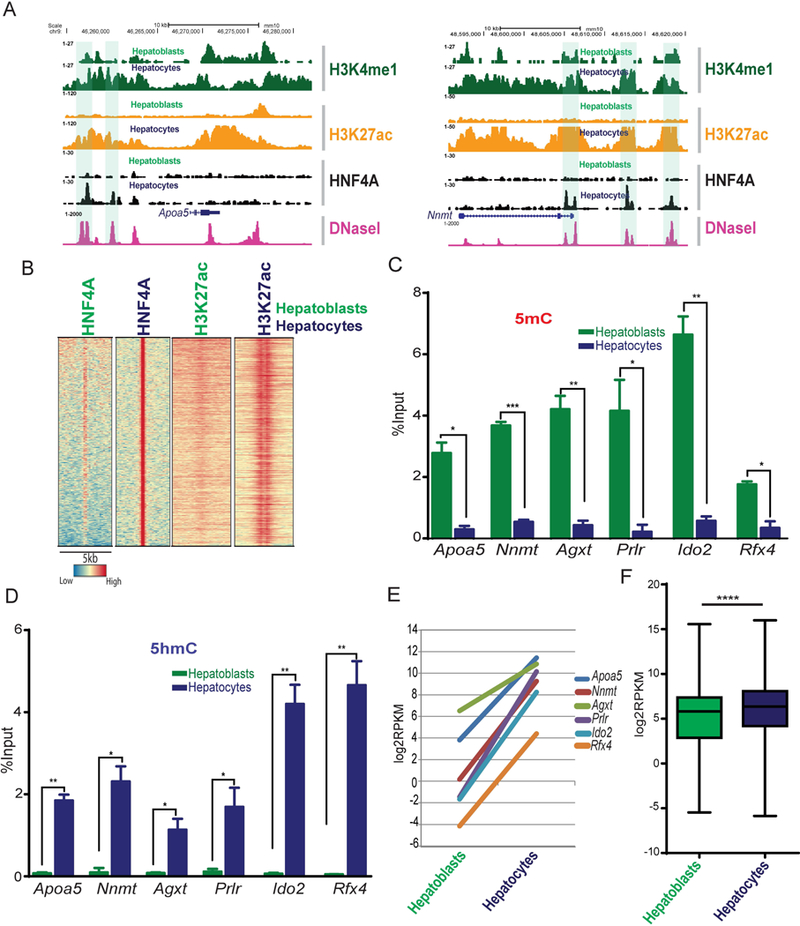
(A) Genome browser view for HNF4A target genes Apoa5 and Nnmt, with ChIP-seq signals for H3K4me1 (dark green), H3K27ac (orange) and HNF4A (black) from hepatoblasts and hepatocytes showing acquisition of HNF4A and increased H3K27ac at hepatocyte-specific HBRs (highlighted). DNase-seq data from the adult liver is shown in pink. (B) Heatmap of HNF4A bound regions that are only significant in hepatocytes, not hepatoblasts. H3K27ac accompanied this differentiation. Colours represent enrichment level. (C) MeDIP-qPCR and (D) hMeDIP-qpcr showing 5mC and 5hmC (respectively) levels at loci that gain HNF4A binding during differentiation from hepatoblasts and hepatocytes (n=3). Values represent mean ±SEM *P<0.05, **P<0.01 by t-test. (E) RNA-seq in hepatoblasts and hepatocytes (log2RPKM values) of genes associated with hepatocyte-specific HNF4A bound regions (Apoa5, Nnmt, Agxt, Prlr, Ido2 and Rfx4). (F) Box plot showing RNA-seq values (Log2RPKM) in hepatoblasts and hepatocytes for all genes associated with hepatocyte-specific HNF4A bound regions.
