Fig. 6. Tet3 transcription is directly regulated by HNF4A.
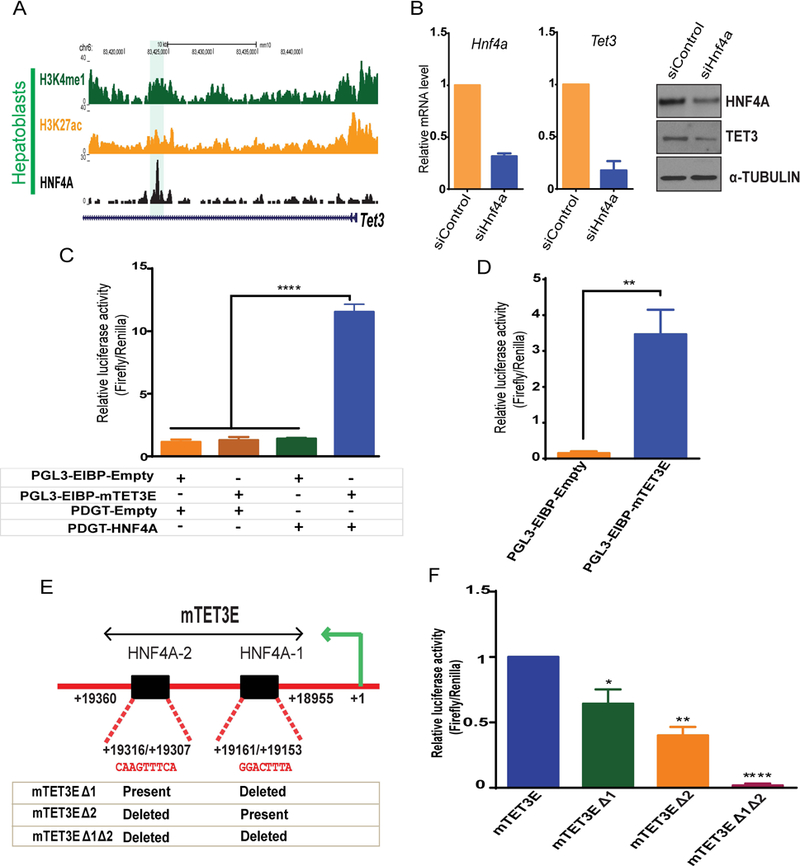
(A) Genomic browser view showing ChIP-seq of HNF4A, H3K4me1 and H3K27ac enrichment at Tet3 locus in hepatoblasts (highlighted). (B) Hnf4a and Tet3 mRNA and protein level in siControl and siHnf4a knockdown in HPPL cells (96 hrs). (C) Luciferase assays using control (PGL3-EIBP-Empty and PDGT-Empty), HNF4A overexpression (PDGT-HNF4A) and the mouse Tet3 enhancer (PGL3EIBP-mTet3E) constructs. Constructs were overexpressed in HEK293T cells in combinations shown below the graph (+ transfected and – not transfected). (D) Luciferase assays in HepG2 cells transfected with control (PGL3-EIBP-empty) or Tet3 enhancer luciferase constructs (PGL3EIBP-mTET3E). (E) Schematic representation of the Tet3 enhancer region (mTet3E) cloned for luciferase assays. Sequence alignments show two HNF4A motifs (HNFA-1 and HNF4A-2) within the Tet3 enhancer. The table represents the constructs generated by deleting HNF4A motifs. (F) Luciferase assays in HepG2 cells transfected with mTET3E or mTET3E carrying one deleted HNF4A motif (mTET3EΔ1 or mTET3EΔ2) or mTET3E with both HNF4A motifs deleted (mTET3Δ1Δ2). Values represent the mean ± SEM. p*<0.05, **p<0.01, ****p<0.0001 from t-test.
