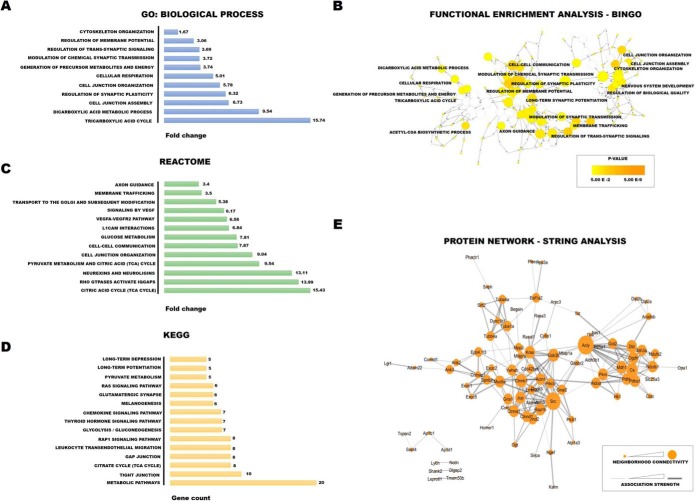
Fig. 9.
Bioinformatics tools for the analysis of proteins with aberrant S-PALM/S-NO crosstalk. A–C, Panther software analysis in terms of (A) biological processes (GO_BP), (B) REACTOME pathway analysis, (C) KEGG pathway analysis (D) STRING analysis of protein interactome. E, Bingo analysis of enriched biological classes. The analysis was done using the 'hyper geometric test', and all GO terms that were significant with p < 0.05 (after correcting multiple term testing by Benjamini and Hochberg false discovery rate corrections) were selected as overrepresented.