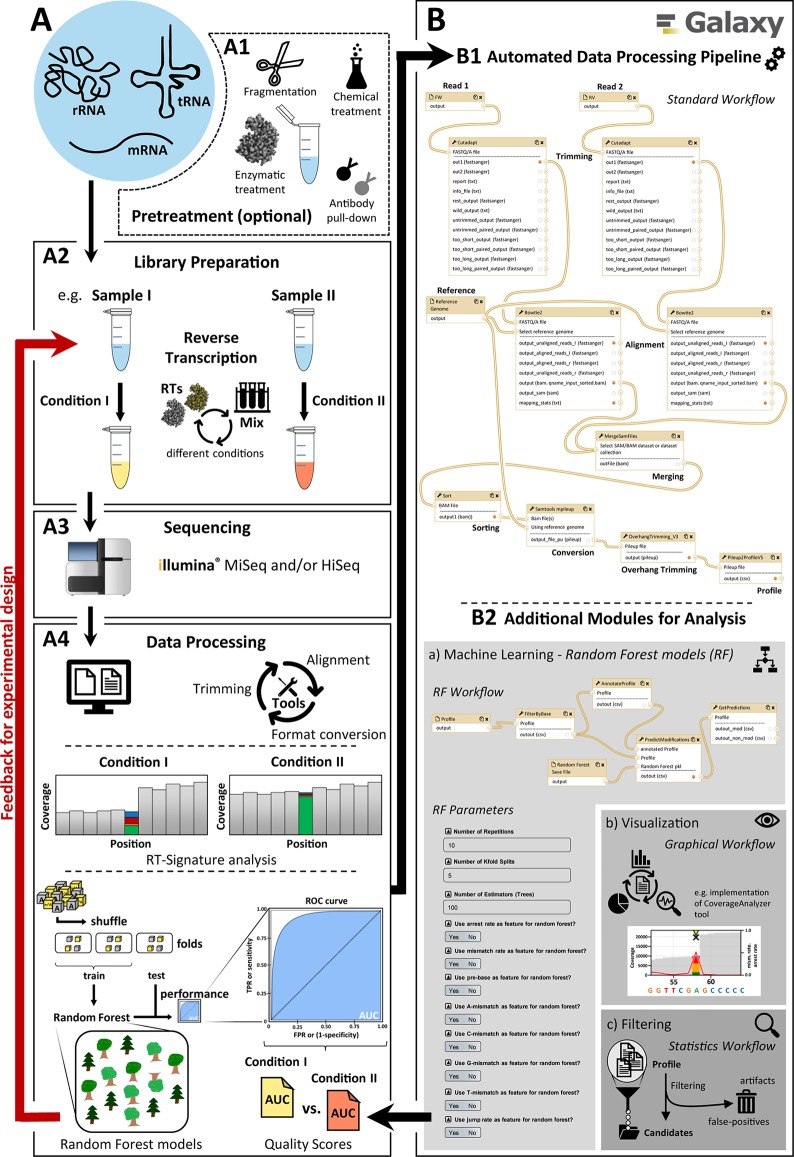
Figure 1.
Main overview of the modification calling pipeline. A diagram showing the different steps for creating and analyzing RNA-Seq data. The pipeline has two parts: (A) general workflow for the processing of RNA samples and (B) the implemented automated graphical workflow system with the available modules for bioinformatics data analysis. (A) consists of (A1) possible and partly necessary pretreatments for different RNA species, (A2) library preparation with the possibility of adaptations (e.g. conditions for reverse transcription), (A3) sequencing with Illumina sequencing platforms (e.g. MiSeq/NextSeq and HiSeq), and (A4) data processing including basic data treatment like adapter trimming, alignment, and format conversion, as well as data analysis (e.g. machine learning and RT-signature analysis). The elaborate data processing (A4) was fully automated in (B) by using the open-source Galaxy platform to create and provide a quick and user-friendly feedback mechanism to optimize the experimental design, sample preparation, and data processing. The standard workflow (B1) is supplemented by various additional modules (B2) including workflows for (a) machine learning, (b) visualization, and (c) filtering.