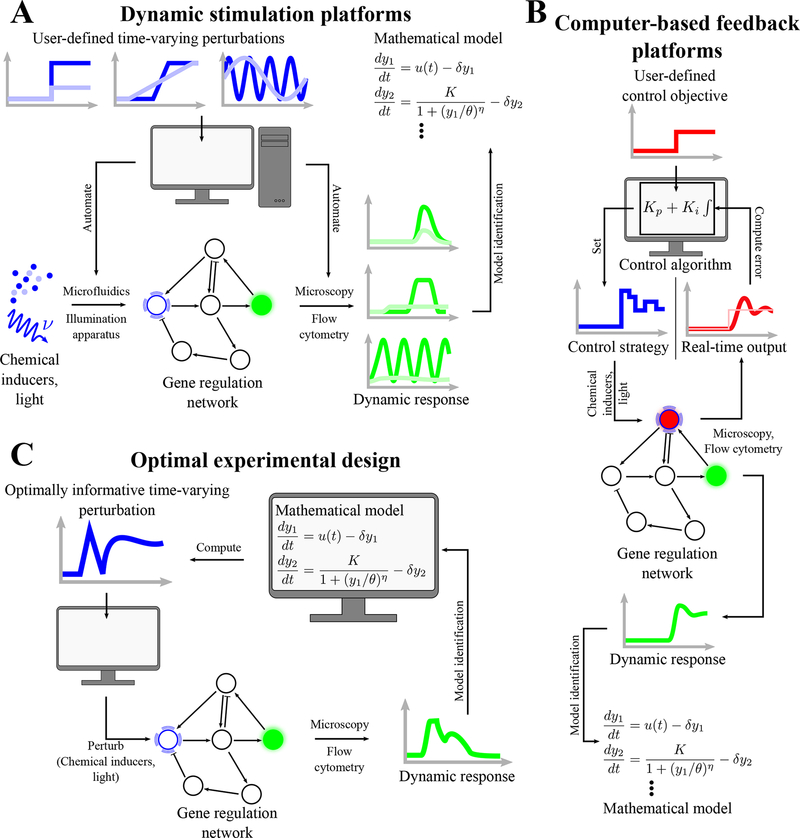
Figure 1.
Computer-based study of gene regulatory networks. (A) Computers are used to subject cells to dynamic stimulations using chemical inducers or light. Automated microscopy or flow cytometry is used to monitor the dynamic response of the gene regulatory network in real time, and the data is then analyzed to infer a mathematical model capturing the dynamics of the system. (B) Automated microscopy or flow cytometry can also be used to implement feedback control through the computer. The state of one or more nodes of the regulatory network is measured and compared to a reference objective that the experimenter chooses a priori. This error is fed into a control algorithm that will modify the chemical or light input in real-time to steer the controlled gene or genes towards the objective. One or more other nodes can also be monitored to study their reaction to this internal perturbation. (C) A model of the system can be used to infer what dynamic perturbation would yield the most information to improve understanding and modeling of the system. The process can be iterated, with the improved model being used to design new perturbations.