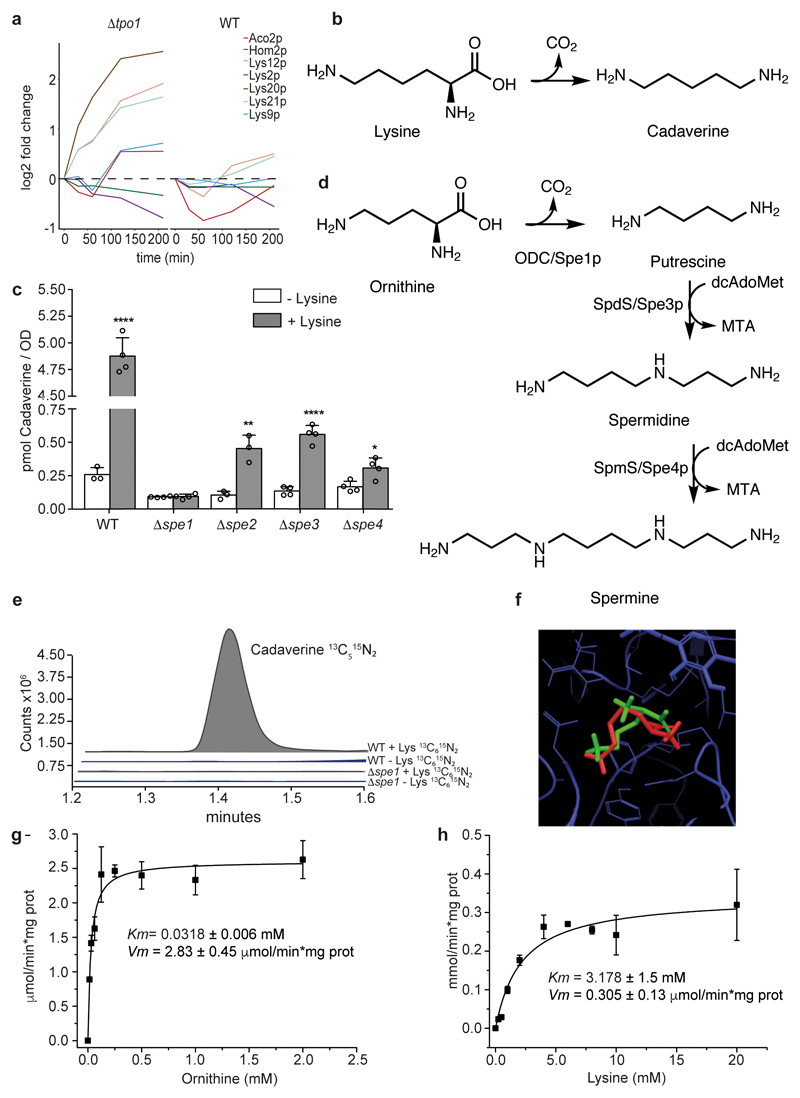
Figure 1. A promiscuous reaction of Spe1p forms cadaverine from lysine in yeast.
a. Lysine biosynthetic enzymes are differentially induced upon exposure to 1.5 mM H2O2 and deletion of TPO1 (Δtpo1). Shown is a 210-min times series 6 recorded by SWATH-MS and re-processed with Spectronaut Pulsar X. Expression levels are shown as log2 (differentially expressed enzymes). b, The decarboxylation of lysine leads to the formation of cadaverine c, Cadaverine concentrations in overnight wild-type (WT) and Δspe1-Δspe4 yeast cultures with or without lysine (25 μg ml-1) supplementation. Mean ± s.d., n = 4 biologically independent samples except for wild-type non-supplemented and Δspe2 supplemented, for which n = 3. Unpaired two-tailed Student’s t-test versus the non-supplemented control: *P = 0.0163, **P = 0.0045, ****P ≤ 0.0001. d, Schematic illustration of the canonical yeast polyamine biosynthesis pathway. ODC, ornithine decasboxylase e, 13C5 15N2 cadaverine is formed from isotope-labelled lysine ([13C6 15N2] lysine) after a 1-h incubation of wild-type strains, but is not formed in Δspe1 cells. This experiment was repeated three times with similar results. f, Results of docking lysine (red) and ornithine (green) to the same site of Spe1p using Glide from Schrödinger suite (v. 2015-3) and a homology model. g, h, In vitro activity of Spe1p on metabolizing ornithine (g) and lysine (h) (mean ± s.d.; n = 3 independent experiments). The saturation curves were fitted to a Michaelis-Menten equation.