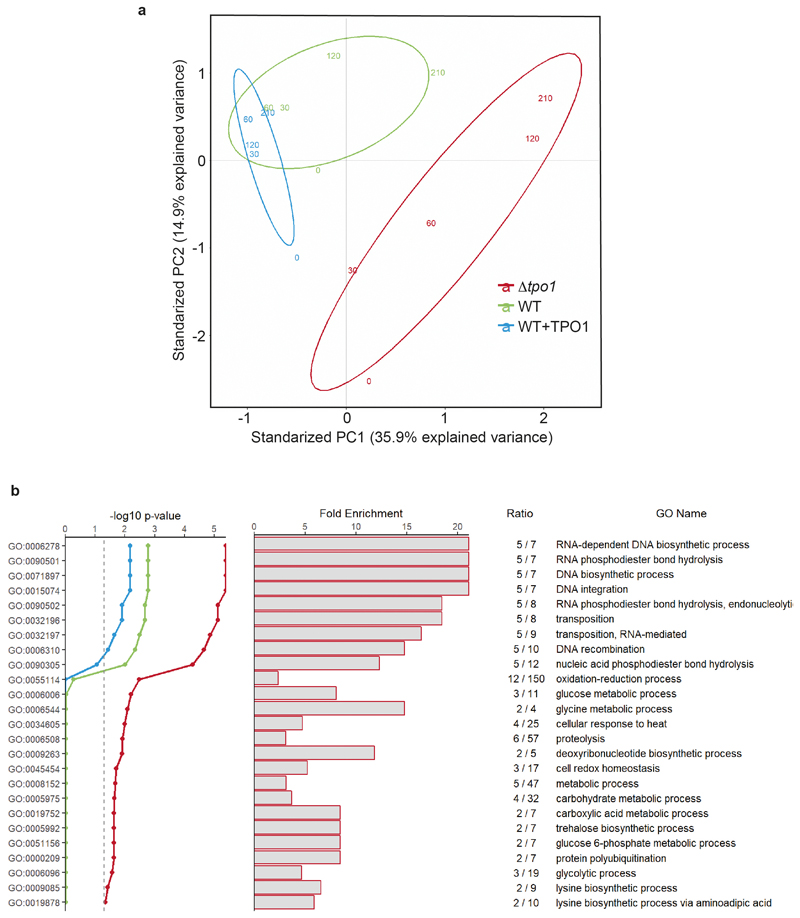
Extended data Figure 1. Proteomic analysis.
A proteomic time-course experiment recorded by SWATH-MS 6, was re-analysed with a new spectral library and a recent version of Spectronaut (Biognosys) software, increasing the depth of the proteomic analysis. a, Principal component analysis shows that proteome profiles of H2O2-exposed Δtpo1 yeast 6 are more different from the wild type than that of a Tpo1p-overexpressing strain and that there is a metabolic adaptation in the lysine pathway. Numbers indicate time points (in minutes) after the oxidative insult. b, Gene Ontology (GO) enrichment analysis shows that a number of biosynthetic processes are significantly enriched in proteins differentially expressed between wild-type and Δtpo1 yeast upon exposure to H2O2. Most of the GO terms that are significantly enriched belong to processes that are typically seen to be activated upon treatment with H2O2, including oxidation-reduction process, cell cycle, nucleotide synthesis and ribosome 43. Metabolic processes affected include carbohydrate- and amino acid metabolism, including two GO terms specific for the lysine pathway. Significance of enrichment over base frequency was calculated using a binominal test 44, and P values (red line) were corrected for false discovery rate using Bonferroni (red) or Benjamini-Hochberg (green) correction. n = 5.