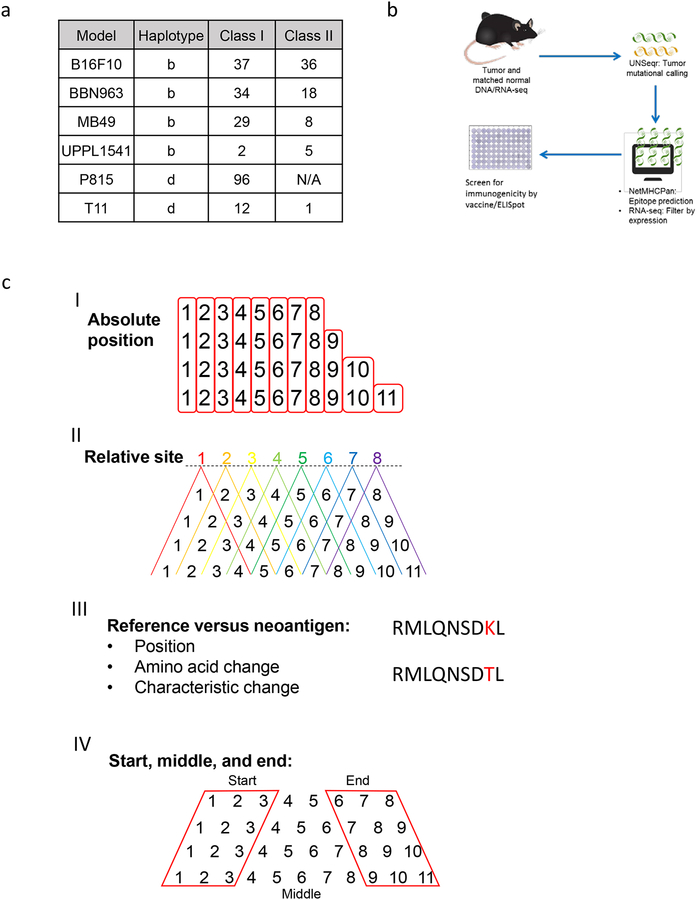
Figure 1: Summary of tumor antigen prediction and identification of peptide-intrinsic features.
(A) Number of MHC class I and II neoantigens/mHA per tumor model contained within the study. (B) Schematic of neoantigen/mHA prediction and ELISpot validation workflow. (C) Summary of the major classes of peptide-intrinsic features identified for each antigen, including amino acid sequence and characteristics at I) each absolute position, II) each relative site, III) the mutation position, and IV) the start, middle, and end of each peptide. Red boxes around columns in I) demonstrate each absolute position and in IV) demonstrate the start, middle, and end distinctions. Red lettering in III) provides an example for SNV mutation site between reference and antigenic sequences.