Figure 2: IDH1 edited clones retain G-CIMP methylation patterns at several G-CIMP defining sites but undergo greater genome-wide loss of methylation than matched IDH1R132H/WT clones.
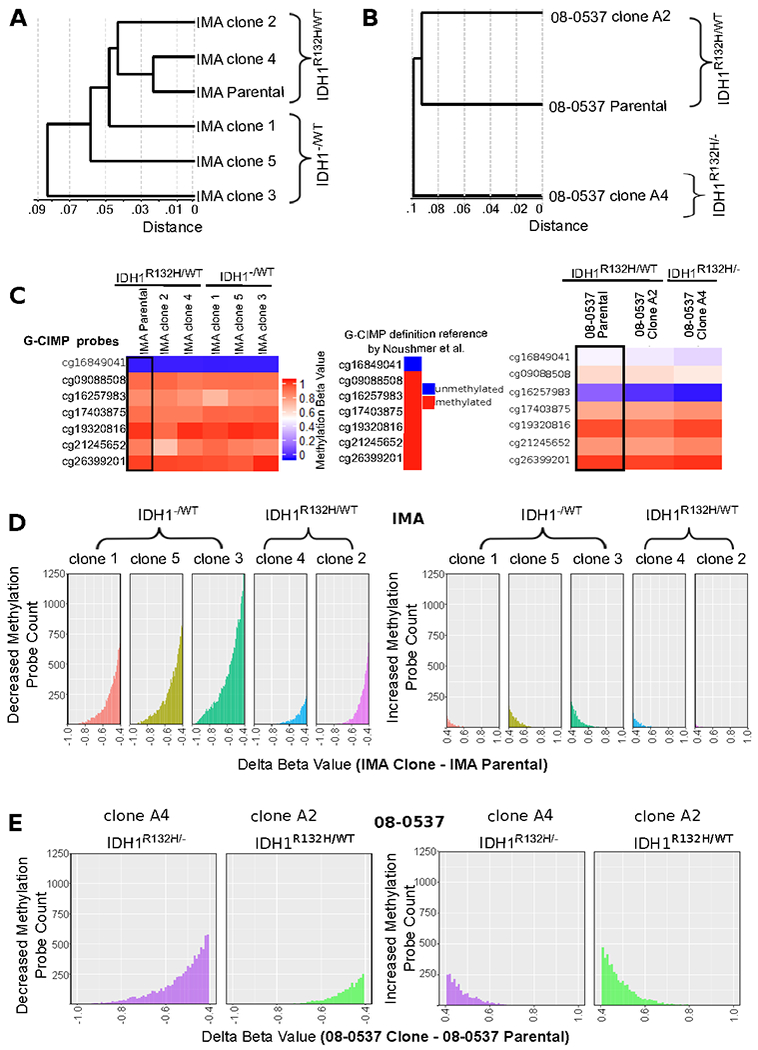
A, Unsupervised hierarchical clustering of IMA clones based on correlation of methylation beta values clusters IMA clones with IDH1R132H/WT genotype with parental IDH1R132H/WT IMA cells. Distance is given in 1-r (pearson correlation distance). B, Similarly, unsupervised hierarchical clustering of 08-0537 clones using the same methodology shows unedited clones cluster with parental cells. C, Observation that G-CIMP sites most indicative of G-CIMP retain G-CIMP linked alterations in methylation pattern in all clones irrespective of IDH1R132H allele status. Note that while 8 sites are typically used for G-CIMP identification on the 450K array only 7 of these sites are present in the methylation EPIC array used in this work. Therefore, conservatively 6/7 sites must display the G-CIMP pattern to enable a call of G-CIMP positive. Site cg16849041 is hypomethylated in G-CIMP and hypomethylated in all IMA samples analyzed. The other 6 sites are hypermethylated in G-CIMP and show hypermethylation (β≥.7) in all IMA clones analyzed. The 08-0537 cells may not fit the definition of G-CIMP positivity based on the 7 site definition; however, sites that are hypermethylated retain hypermethylation in all clones. D, Histogram plots of genome-wide beta value differences (Parental IMA cells – averaged values from either IDH1R132H/WT clones or IDH1−/WT clones) shows that methylation loss is the predominant change in methylation identified in the genome-wide pattern of clones; and, the IDH1−/WT cells show greater loss of genome-wide methylation compared to IDH1R132H/WT clones. Each clone was passaged at least 10 population doublings post-single cell cloning. E, The 08-0537 clone with loss of heterozygosity at the IDH1 locus also shows more pronounced CpG demethylation patterns than its matched unedited clone.
