Figure 3: Demethylation is observed in a subset of the 131 CpG probes originally annotated as hypomethylated in G-CIMP-low.
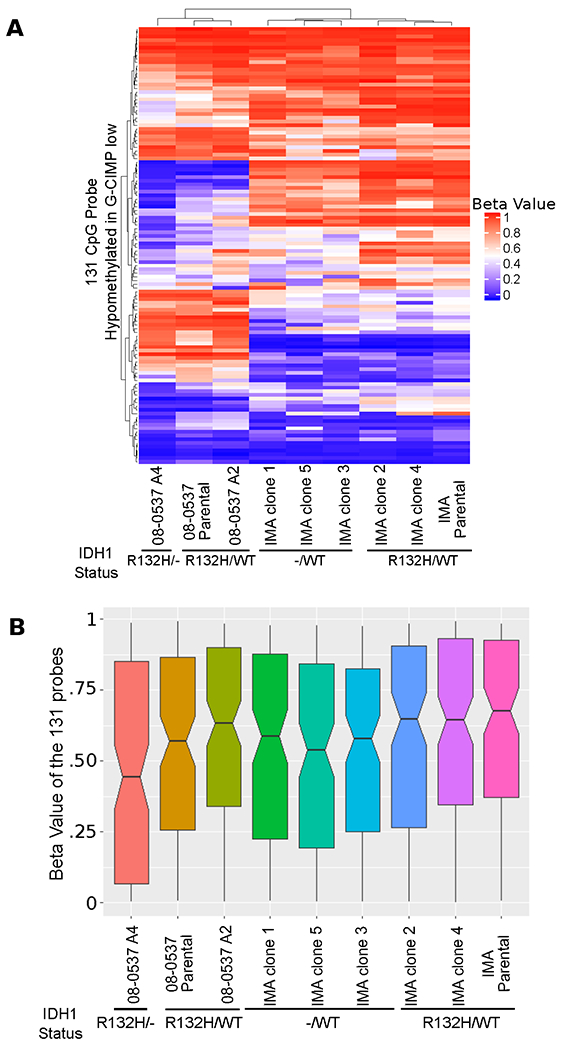
A, Unsupervised hierarchical clustering based on Euclidean distance of 131 probes hypomethylated in G-CIMP-low shows that, within cell lines, cells with intact heterozygous IDH1R132H/WT mutations tend to cluster together, and this clustering is likely driven by demethylation of a subset of probes in clones with altered IDH1. B, Notched boxplots showing median and confidence interval for methylation level of the 131 CpG probes. Clones with altered IDH1 show the largest decreases in median methylation level compared to their matched unedited controls.
