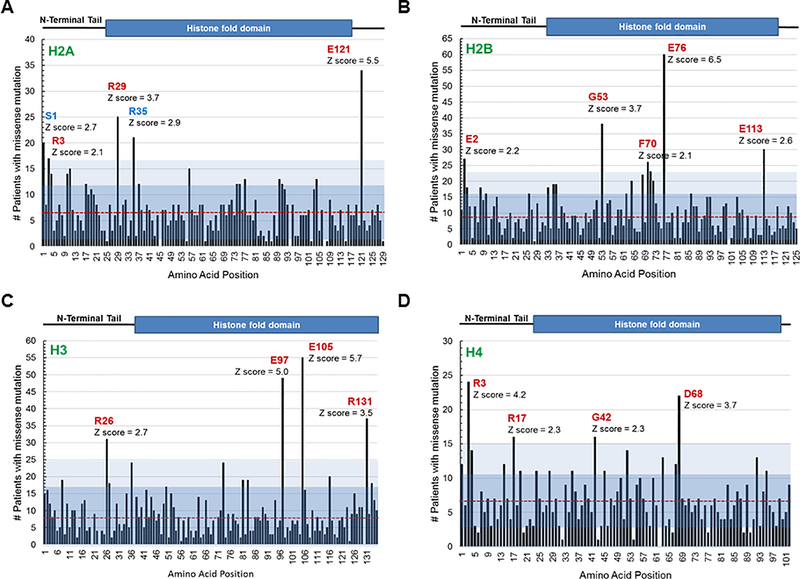
Figure 1. Survey of the most frequently found canonical histone mutations in cancer patient samples.
A cross cancer mutation summary was performed using the cBioPortal to search a total of 41,738 non-redundant patient samples across all cancer types. The number of patients reported to have a missense mutation at each amino acid residue position across histone paralogs was graphed. A red line denotes the average number of mutations for each histone across all paralogs, and blue shaded regions indicate the first two standard deviations from the average. Location of the cumulative missense mutations found in: (A) 15 canonical H2A genes (HIST1H2AA/B/C/D/E/G/H/I/J/K/L/M, HIST2H2AB/C, HIST3H2A), (B) 18 canonical H2B genes (HIST1H2BA/B/C/D/E/F/G/H/I/J/K/L/M/N/O, HIST2H2BE/F, HIST3H2BB), (C) 12 canonical H3 genes (HIST1H3A/B/C/D/E/F/G/H/I/J, HIST2H3D, HIST3H3), and (D) 14 canonical H4 genes (HIST1H4A/B/C/D/E/F/G/H/I/J/K/L, HIST2H4A, HIST4H4) across all cancer patient samples. The Z scores indicate the most frequently found mutations (Z > 2) in each histone. Amino acids displayed in red were significantly more frequent in cBioPortal than the general SNP database.