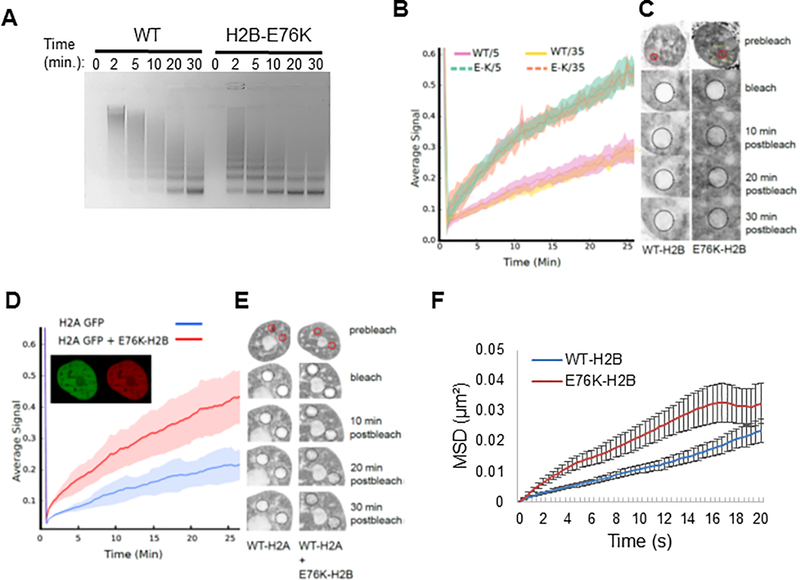
Figure 6. H2B-E76K fundamentally alters chromatin structure and dynamics.
(A) Micrococcal nuclease (MNase) assay with nuclei of MCF10A cells stably expressing either WT or H2B-E76K demonstrate that E76K expression significantly increases sensitivity to MNase. Digest efficiency was visualized by agarose gel. (B) Fluorescence recovery after photobleaching (FRAP) analysis demonstrates that H2B-E76K has significantly faster chromatin dynamics than WT. FRAP analysis was carried out after induction of either WT or E76K mutant H2B-GFP fusions for 5 or 35 days in MCF10A cells. Dashed lines represent cells expressing H2B-E76K, solid lines represent data from cells expressing inducible GFP-tagged WT H2B (n = 10 cells). (C) Representative FRAP assay pre-bleach, bleach and post-bleach images of nuclei expressing GFP-tagged WT or H2B-E76K indicate faster fluorescent recovery in cells expressing E76K. (D) FRAP analysis of histone H2A-GFP dynamics in MCF10A cells expressing only H2A-GFP (n = 20 cells) or both H2A-GFP and a mutant H2B E76K mCherry fusion (n = 25 cells) with standard deviation envelopes. Inset demonstrates co-expression of H2A GFP and E76K mCherry. (E) Representative pre-bleach, bleach and post-bleach images of H2A GFP in WT MCF10A cells or in cells co-expressing mCherry tagged H2B-E76K. (F) 100nm particles injected into the nucleus had significantly increased mean square displacement (MSD) over time in MCF10A cells stably expressing exogenous H2B-E76K (red) compared to cells expressing WT H2B (blue). N=68 (WT) and N=67 (E76K) cells were analyzed. Error bars represent SEM.