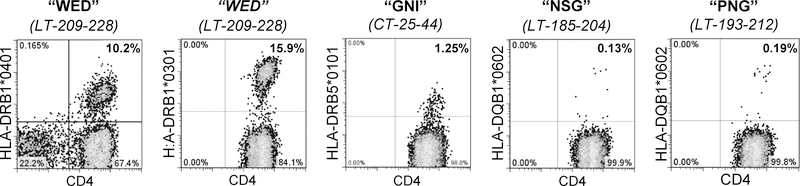
Figure 1: TGEM identified four peptides presented by four population-prevalent HLA class II alleles.
TGEM was performed as outlined in the Methods testing seven allele types. PBMCs were cultured in the presence of pools of five MCPyV 20-mer peptides (WED, GNI, NSG, PNG; amino acid sequence indicated) for 10–14 days, followed by staining for pools of tetrameric complexes of 10 peptides (composed of two combined pools) bound to HLA class II molecules. After identification of CD4+tetramer+ T cells, the same cultures were stained with single tetramers containing each relevant peptide within the positive pool. Representative flow plots are shown for each peptide. For each epitope, four individuals were tested, and one to four persons were positive as detailed in column 7 of Table 1. The percentage of viable CD4+tetramer+ T cells (negative for CD8/CD14/CD19) in the lymphocyte forward/side scatter region are denoted.