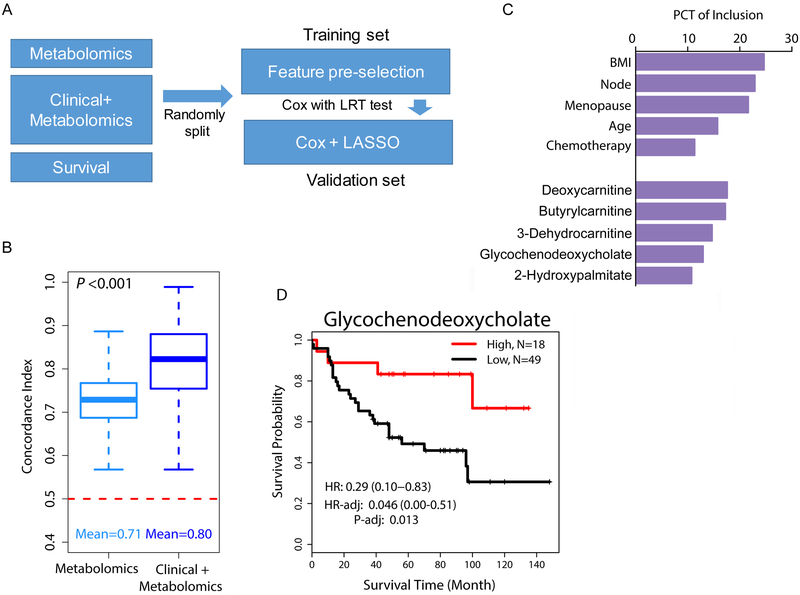
Figure 1. Metabolomics improves survival prediction for breast cancer.
A. Computational approach to build a survival prediction model using LASSO feature selection for Cox proportional hazards modeling. B. Concordance indexes for two models, either including only metabolomics data (398 metabolites) or a combination of clinical and metabolomics data (clinical + metabolomics). The dashed red line (C-index = 0.5) would represent models with no prognostic power whereas a C-index of 1 indicates a perfect prediction accuracy. Shown are the medians and confidence intervals not crossing the red line. C. Top ten features selected from the training set using LASSO feature selection for survival prediction in the validation set. PCT, percentage. D. Association of tumor glycochenodeoxycholate content with breast cancer-specific survival. Kaplan-Meier plot and hazard ratio (HR) estimates in the unadjusted and adjusted Cox regression analysis. High: elevated glycochenodeoxycholate levels (n=18). Low: detection limit or below (n=49). Adjusted analysis with patient’s age, BMI, menopausal and node status, and receipt of chemotherapy as covariables.