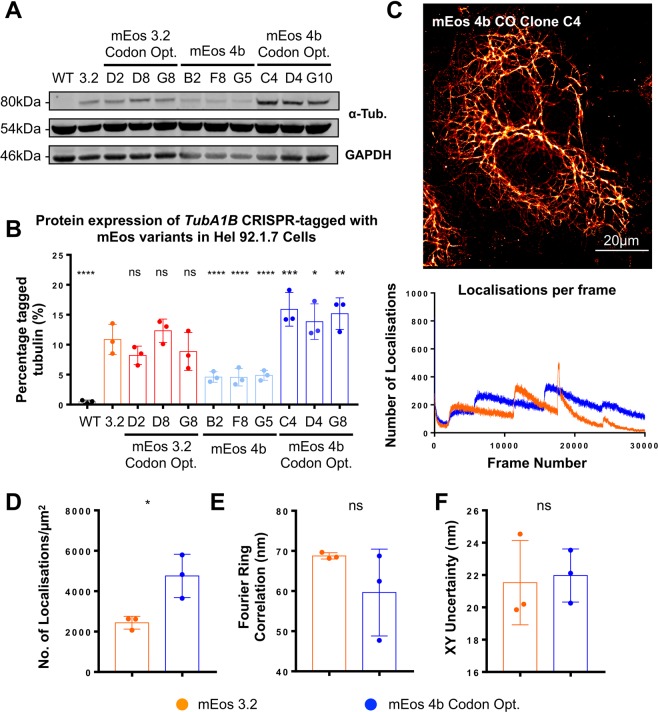
Figure 2.
Optimised expression of mEos significantly improves SMLM imaging. Hel 92.1.7 clones CRISPR edited to express mEos 3.2, codon optimised mEos 3.2, mEos 4b, and codon optimised mEos 4b TubA1B inserts were isolated and compared to a previously reported TubA1B-mEos 3.2 clone to determine the effects of fluorophore properties and codon optimisation on expression level. (A) Western blotting of clones carrying specific mEos variants at the TubA1B locus showed a variation in expression of tagged tubulin dependent on the insert. (B) No significant difference was observed in mEos 3.2 codon optimised clones (ns p = 2.189, ns p = 0.2465, ns p = 0.1178), however a significant reduction in expression was observed in cells carrying mEos 4b when compared to the mEos 3.2 clone (****p < 0.0001, ****p < 0.0001, ****p < 0.0001). Conversely, a significant improvement in expression was observed in clones carrying a codon optimised mEos 4b (***p = 0.0005, *p = 0.0244, **p = 0.0022). (C,D) mEos 4b codon optimised (CO) clones produce high quality PALM images, with a significantly increased number of localisations compared to mEos 3.2 clones (*p = 0.0230). (E,F) No significant difference in fourier ring correlation or localisation precision were observed (ns p = 0.2819, ns p = 0.8161). (n = 3 (S.D), statistical analysis performed using One-Way ANOVA with multiple comparisons to the mEos 3.2 clone for western data, and by t-test for quantitative SMLM comparisons. Complete unedited gels are included in Fig. S2).