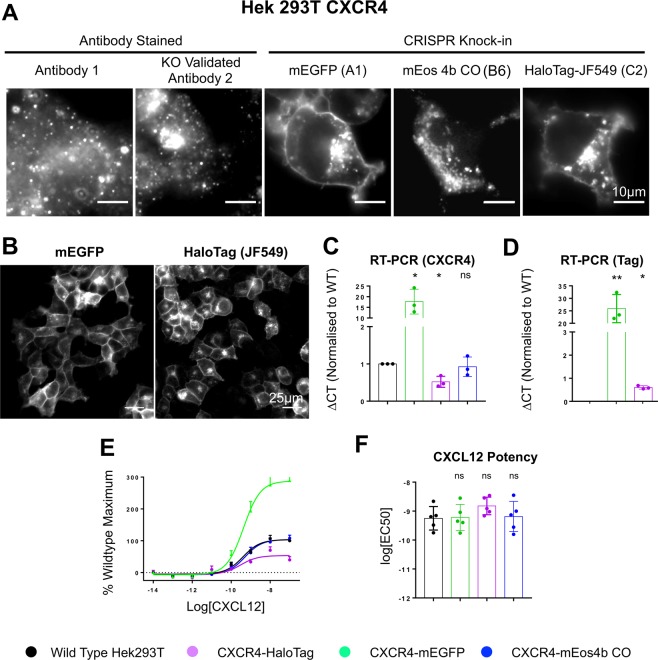
Figure 4.
CRISPR-Cas9 mediated knock-in at the CXCR4 C-terminus results in endogenously labelled protein which localises to the membrane and endosomes. (A) Staining of CXCR4 with anti-CXCR4 antibody 1 or KO validated antibody 2 results in poor labelling of the receptor protein, while mEGFP and HaloTag knock-in cells demonstrate a membrane specific localisation. Interestingly mEos 4b CO clones demonstrate fluorescent aggregates, suggesting the fluorophore is cleaved. (B) mEGFP and HaloTag knock-in samples provide clonal populations which are evenly labelled and offer large samples for imaging experiments. (C) Measurement of total CXCR4 expression in wild type versus CRISPR edited cells through qRT-PCR. Compared to wild type, mEGFP knock-ins demonstrate an approximately 20-fold increase in expression (*p = 0.0384). Conversely, HaloTag knock-ins demonstrate a 50% reduction in expression (*p = 0.0301), while mEos 4b CO knock-in shows no change (ns p = 0.6661). (D) When RT PCR is performed using tag specific primers, the significant increase in mEGFP expression is likely due to the stability of mEGFP tagged CXCR4 RNA, suggesting that RNA stability is likely a factor in mediating the tag specific down-regulation observed (**p = 0.0050, *p = 0.0155). (E) CXCL12-mediated G protein activation measured by observing G protein dissociation/conformational changes by BRET in knock-in or wildtype cells transfected with cDNA encoding Gαi1/Nluc and Venus/Gγ2. Results are expressed as % maximal CXCL12 response observed in wildtype cells andshow a 3 fold increase in response in mEGFP knock-in clones compared to wild type, with a 50% reduction in response in HaloTag knock-in, and no variation in mEos 4b CO clones. (F) Potency of CXCL12-mediated G protein activation in cells expressing wildtype or genome-edited CXCR4 (ns p = 0.0549, ns p = 0.9736, ns p = 0.5770). (n = 3 (S.D) for qRT-PCR data, compared by One-Way ANOVA with multiple comparisons. n = 5 for functional data).