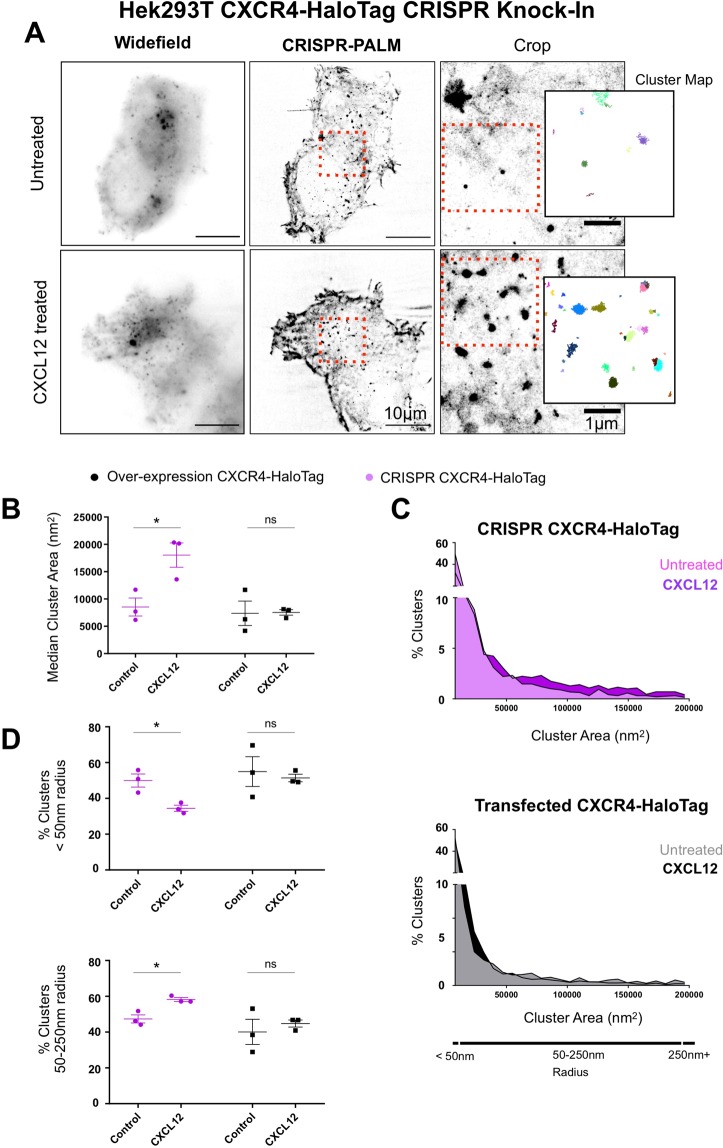
Figure 5.
CRISPR Knock-In CXCR4 clones allow for accurate single molecule cluster analysis compared to transfected cells. (A) Single molecule imaging of HaloTag CXCR4 Knock-Ins allows for effective single molecule localisation microscopy through labelling with Janelia Fluor 549, and subsequent cluster analysis using persistence based clustering. (B) CXCR4 knock-in and Hek293T transfected with a CXCR4-HaloTag were subject to single molecule imaging and cluster analysis after treatment with CXCL12 (10μM). CRISPR knock-ins demonstrate a significant increase in median cluster area on ligand treatment (*p = 0.0272), while transfected cells show no significant change. (C) Histograms showing percentage of clusters across a range of areas show that while a substantial shift towards larger cluster sizes is evident in CRISPR knock-ins, this is reduced in transfected cells. (D) The percentage of clusters below 50 nm in radius is significantly reduced in CRISPR cells (*p = 0.0384) upon treatment with CXCL12 (upper panel), with a parallel increase in the percentage of clusters of larger sizes consistent with endocytic trafficking observed (*p = 0.0133) (lower panel). Conversely, no significant change in either collection of cluster sizes is evident in transfected cells. (n = 3, S.E.M, paired t-test).