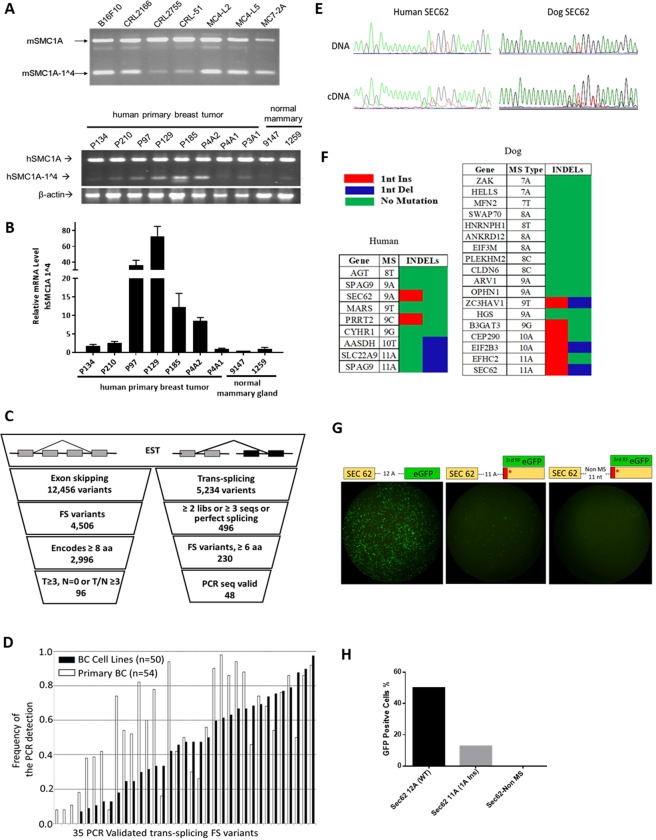
Figure 2.
Detecting FS variants. (A) End-point RT-PCR analysis of the mSMC1A-1^4 in mouse tumor cell lines and human hSMC1A-1^4. (B) End-point RT-PCR analysis and RT-qPCR of the human hSMC1A-1^4 expression in human primary breast tumor tissues and normal mammary tissues. All values are normalized relative to the expression levels in sample 1259 (set as 1). Data are mean 2−ΔΔC of triplicates with SD. (C) Analysis of the human EST database for FS variants by exon skipping and trans-splicing. (D) Analysis of the frequency of the expression of the 35 trans-splicing variants in 50 human breast cancer cell lines and 54 primary human breast tumors. (E) Example of a sequence trace of the MS region in SEC62 dog and human genes in paired DNA/cDNA samples. (F) Summary sequencing results of microsatellite candidates in human (4 breast cancer cell lines) and dog (primary dog tumor tissues). (G) Ex vivo analysis of the MS INDEL in transcription and translation of the MS INDEL variants. eGFP was fused to the 3rd reading frame after 11 A MS of SEC62 or after 11 non-MS nucleotides. The eGFP directly fused to 12 A MS was the positive control. The three different plasmids were transfected individually into 293 T cells and GFP fluorescence was measured 24 hrs after transfection. (H) FACS analysis of the GFP positive cells.