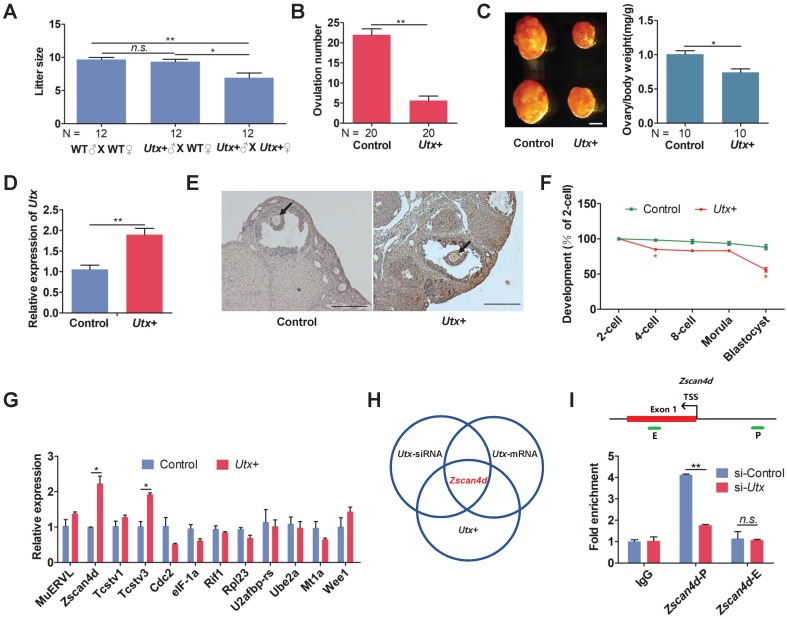
Figure 3.
Utx transgenic mice also exhibit ZGA defects. (A) Numbers of pups per litter. ♂, ♀, and X indicate the male, female, and cross mate, respectively. WT, wild-type; N, total number of couples; n.s., not significant. Error bars indicate SEM. *P < 0.05, **P < 0.01 by the two-tailed Student's t-test. (B) Ovulation number of Utx+ females following hCG injection. N, total number of females. Error bars indicate SEM. **P < 0.01 by the two-tailed Student's t-test. (C) Images representative of the control and Utx+ mice ovary (left), bar graphs showing the ratio of ovary/body weight (right). N, total number of females. Error bars indicate SEM. *P < 0.05 by the two-tailed Student's t-test. Scale bar, 500 μm. (D) qPCR results showing mRNA levels of Utx in Utx+ mice 2-cell embryos. Error bars indicate SEM. Values were normalized to Gapdh. **P < 0.01 by the two-tailed Student's t-test. (E) Myc immunohistochemistry staining in wild-type (WT) and Utx+ mice on ovaries prepared from 8-week-old mice. Black arrows indicate oocytes. Scale bar, 200 μm. (F) Developmental rates in the Utx+ mice embryos cultured in vitro. The efficiency was calculated based on the number of 2-cell embryos. Error bars indicate SD, n ≥ 3. *P < 0.05 by the two-tailed Student's t-test. (G) qPCR results showing mRNA levels of ZGA markers in Utx+ mice 2-cell embryos. Error bars indicate SEM. All values were normalized to Gapdh. *P < 0.05 by the two-tailed Student's t-test. (H) Schematic illustration of the target gene of UTX. (I) Location of PCR amplicons in the Zscan4d gene used for ChIP analyses (upper). ChIP-qPCR assays with an anti-UTX antibody at the indicated Zscan4d locus in si-control and si-Utx-injected 2-cell embryos (Zscan4d-E: exon 1 region, Zscan4d-P: promoter region) (bottom). Error bars indicate SEM. IgG served as the control. **P < 0.01 by the two-tailed Student's t-test. E, Exon 1; P, promoter; TSS, transcription start site; n.s., not significant